| PHILOSOPHIE | HISTORY OF BIOLOGY | PHILOSOPHY |
|
|||||||
|
|
|
|
|
Thèmes
|
|
|
Themata
|
|
BioSapiens
|
||
|
EuroPathoGenomics
|
||
| Constraints on the cell chassis | ||
|
|
||
![]() This sign indicates that the corresponding work
has been performed with the HKU-Pasteur
Research Centre, Hong Kong
This sign indicates that the corresponding work
has been performed with the HKU-Pasteur
Research Centre, Hong Kong
Epigenesis: cognition, immunology and epidemiology
|
|
JP Changeux, P
Courrège, A Danchin A theory of the epigenesis of neuronal networks by selective stabilization of synapses Proc Natl Acad Sci U S A (1973) 70: 2974-2978 |
JP Changeux, A
Danchin
Selective stabilisation of developing synapses as a mechanism for the
specification of neuronal networks
Nature (1976) 264: 705-712
|
|
|
A Danchin
A selective theory for the epigenetic specification of the monospecific
antibody production in single cell lines
Ann Immunol (Paris) (1976) 127: 787-804
|
|
|
![]() A Danchin
A Danchin
Stabilisation fonctionnelle et épigenèse: une approche biologique de la
genèse de l'identité individuelle
In: "L'Identité" (JM Benoist, ed) Grasset (1977): 185-221
|
|
|
A Danchin
The specification of the immune response: a general selective model
Mol Immunol (1979) 16: 515-526
![]() JP Changeux, P Courrège, A Danchin, JM Lasry
JP Changeux, P Courrège, A Danchin, JM Lasry
Un
mécanisme biochimique pour l'épigenèse de la jonction neuro-musculaire
C R Séances Acad Sci III (1981) 292: 449-453
TW Ng, G Turinici,
A Danchin
A double epidemic model for the SARS propagation
BMC Infect Dis (2003) 3: 19 ![]()
TW Ng, G Turinici, WK
Ching, SK Chung, A Danchin
A parasite vector-host epidemic model for TSE propagation
Medical Science Monitor (2007) 13: 59-66 ![]()
| A Danchin, A Sekowska The role of information in evolutionary genomics of bacteria In: Evolutionary genomics and systems biology (G Caetano-Anollés, Editor), Wiley & sons, Hoboken, NJ (2010) pp 81-94 |
| Back to top |
Philosophy of Synthetic Biology: epistemology, ethics, history of sciences
A Danchin
The tree and the ring.
Hierarchical and acentered structures in biology [a
paper in System's Biology that exposes the function of closure
in creating global behaviour]
The Cancer Journal (1989) 2: 285-287
V de Lorenzo, A Danchin
Synthetic biology: discovering new worlds and new words
EMBO Rep (2008) 9: 822-827 ![]()
![]() A Danchin
A Danchin
Sélection naturelle et immortalité
in: Les mondes darwiniens
(T Heams, P Huneman, G Lecointre, M Silberstein, eds), Syllepse (2009)
pp 445-470 ![]()
Tous les organismes – bactéries incluses – vieillissent et meurent. En raison de l’incroyable diversité de ce processus (les plantes vieillissent lentement, par exemple), c’est un lieu commun des études sur le vieillissement que de considérer qu’il s’agit d’un paradoxe, qui requiert par conséquent une explication en profondeur. Cette observation a suscité une profusion d’études fascinantes (et de discussions enflammées) destinées à mettre au jour la variété des causes et des mécanismes impliqués dans le vieillissement. Dans ce contexte, l’« immortalité » est considérée comme la norme. Pourquoi et comment vieillissons-nous ? Ce n’est pas cette question que je traiterai ici, je vais prendre le chemin exactement opposé. Considérant que le vieillissement est une conséquence directe de la physique – et c’est en effet l’une des conclusions majeures à laquelle parviennent ceux qui analysent le vieillissement en profondeur quand il repensent les théories de l’« avalanche d’erreurs» d’Orgel ou le « cliquet de Muller –, j’observe au contraire que c’est l’immortalité (que nous observons dans le cancer) qui soulève un paradoxe...
A. Danchin
Information of the chassis and information of the program in synthetic
cells
Syst Synth Biol (2009) 3(1-4):125-134 ![]()
![]()
![]()
A Danchin
Perfect time or perfect crime?
EMBO Reports (2010) 11: 74 ![]()
Opening: Imagine the situation: “[O]ne day, you notice an old country woman crossing a downtown street, just about to put one foot down on the rails of the streetcar line. […] Suppose, now, that at the very moment she puts her foot on the rail a streetcar comes rushing down the tracks toward her. If the old woman does not notice the car and continues across the tracks nothing will happen. But if someone should happen to shout ‘Look out, old woman!’ what would be her natural reaction? […] she would suddenly become flustered and would pause to decide whether to go on or step back […] the mere words ‘Look out, old woman!’ would be as dangerous a weapon as any knife or firearm. […] the man who sounds the warning actually becomes a murderer!" (Ranpo, 1956). In this sense, the Asilomar conference on genetic engineering in 1974 was the perfect crime in using cautionary exclamations asa weapon that had far-reaching consequences for the emerging field of molecular biology: a moratorium stopped research in some places while others proceeded and took the lead. The same fate may lie in store for synthetic biology (SB). The usual crooners have understood the importance of resorting to appeals to ethics and playing on people’s fears to make themselves known and have begun a discussion of the purported dangers of creating organisms a la carte. Such clamouring for attention may yet end up being a shot to our own feet. Moreover, does minimization of a bacterial genome really pose ethical questions? Should we worry, and if so why? After all, what is the purpose of SB? We need to explore the knowledge that we are required to master to (re)construct life in order to gain a better idea of the economic and ethical questions that we have to ask before we start asking them.
Danchin A
Artificial intelligence-based prediction of pathogen emergence and
evolution in the world of synthetic biology
Microb biotechnol (2024) 17: e70014 doi:
10.1111/1751-7915.70014
C Médigue, A
Viari, A Hénaut, A Danchin
Escherichia coli molecular genetic map (1500 kbp): update II
Mol Microbiol (1991) 5: 2629-2640
|
|
|
A Hénaut, T
Rouxel, A Gleizes, I Moszer, A Danchin
Uneven distribution of GATC motifs in the Escherichia coli
chromosome, its plasmids and its phages
J Mol Biol (1996) 257: 574-585
|
|
|
A Hénaut, A Danchin
Analysis and Predictions from Escherichia coli sequences or E.
coli in silico
In: Escherichia coli and Salmonella, Cellular and
Molecular Biology (Editor in Chief: Frederick C Neidhardt) vol. 1, chap
114, pp 2047-2065
|
|
|
A Hénaut, F
Lisacek, P Nitschké, I Moszer, A Danchin
Global analysis of genomic texts: the distribution of AGCT
tetranucleotides in the Escherichia coli and Bacillus
subtilis genomes predicts translational frameshifting and
ribosomal hopping in several genes
Electrophoresis (1998) 19: 515-527
|
|
|
EPC Rocha, A
Viari, A Danchin
Oligonucleotide bias in Bacillus subtilis: general trends and
taxonomic comparisons
Nucleic Acids Res (1998) 26: 2971-2980
|
|
|
EPC Rocha, A
Danchin, A Viari
Analysis of long repeats in bacterial genomes reveals alternative
evolutionary mechanisms in Bacillus subtilis and other competent
prokaryotes
Mol Biol Evol (1999) 16: 1219-1230
EPC Rocha, A
Danchin, A Viari
Functional and evolutionary roles of long repeats in prokaryotes
Res Microbiol (1999) 150: 725-733
|
|
|
EPC Rocha, A
Danchin, A Viari
Translation in Bacillus subtilis: roles and trends of initiation
and termination, insights from a genome analysis
Nucleic Acids Res (1999) 27: 3567-3576
|
|
|
EPC Rocha, A
Danchin, A Viari
Universal replication biases in bacteria
Mol Microbiol (1999) 32: 11-16
|
|
|
EPC Rocha, A
Danchin, A Viari
The DB case: pattern matching evidence is not significant
Mol Microbiol (2000) 37: 216-218
|
|
|
|
|
EPC Rocha, A Danchin |
| Ongoing evolution of strand composition in bacterial genomes | |
| Mol Biol Evol (2001) 18: 1789-1799 |
|
|
|
EPC Rocha, A
Danchin, A Viari
Evolutionary role of restriction/modification systems as revealed by
comparative genome analysis
Genome Res (2001) 11: 946-958 ![]()
|
|
EPC Rocha, A Danchin |
| Base composition bias might result from competition for metabolic resources | |
| Trends Genet (2002) 18:
291-294 |
|
|
X Xia, T Wei, Z Xie, A Danchin |
| Genomic changes in nucleotide and dinucleotide frequencies in Pasteurella multocida cultured under high temperature | |
| Genetics (2002) 161: 1385-1394 |
|
|
Y Wang, EPC Rocha, FCC Leung, A Danchin |
| Cytosine methylation is not the major factor inducing CpG dinucleotide deficiency in bacterial genomes | |
| J Mol Evol (2004) 58: 692-700 |
|
|
|
|
|
E Larsabal, A Danchin |
| Genomes are covered with ubiquitous 11bp periodic patterns, the "class A flexible patterns" | |
| BMC Bioinformatics (2005) 6:
206 |
|
|
|
|
|
G Pascal, C Médigue, A Danchin |
| Universal biases in protein composition of model prokaryotes | |
| Proteins (2005) 60: 27-35
|
|
|
|
G Pascal, C
Médigue, A Danchin
Persistent biases in the amino-acid composition of prokaryotic proteins
Bioessays (2006) 28: 726-738 ![]()
![]()
![]()
| Back to top |
The principles of molecular biology and their formal consequences
![]() A Danchin
A Danchin
Règles de réécriture en biologie moléculaire
Change (1974) 19: 10-24
|
|
|
A Danchin
Genome diversity: A grammar of microbial genomes
ComPlexUs (2004/2005) 2: 61-70 ![]()
![]()
G Fang,
EPC Rocha, A Danchin
How essential are non-essential genes?
Mol Biol Evol (2005) 22: 2147-2156
![]()
|
|
|
M
Bailly-Bechet, A Danchin, M Iqbal, M Marsili, M Vergassola
Codon usage domains over bacterial chromosomes
PLoS Computational Biology (2006) 2: e37 ![]()
![]()
![]()
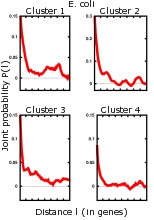 ( 20 K)
( 20 K) |
An issue left unexplained in this paper is the striking quantitative difference between E. coli and B. subtilis. This is clearly visible in Fig. 6, where it is shown the probability that two genes at distance l belong to the same cluster of codon usage. Clusters are characterized by a similar codon bias and were identified using a novel information-based clustering method. While both curves decay on distances sizably longer than what could be accounted by operons, B. subtilis curve manifestly features much longer correlations. It is hard to develop a biologically well-founded explanation for such a striking difference between the two organisms. |
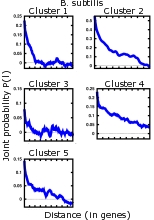 (28 K)
(28 K) |
|
|
|
A Danchin
Archives or palimpsests? Bacterial genomes unveil a scenario for the
origin of life
Biological Theory
(2007) 2: 52-61 ![]()
![]() A Danchin
A Danchin
De la biologie symplectique à la biologie synthétique
In: L'Homme Artificiel (sous la direction de JP Changeux), Odile
Jacob, Paris (2007) pp 61-86
A Danchin, G
Fang, S Noria
The extant core bacterial proteome is an archive of the origin of life
Proteomics (2007) 7: 875-889 ![]()
![]()
![]()
![]()
G Fang, EP Rocha, A Danchin
Persistence drives gene clustering in bacterial genomes
BMC Genomics (2008) 9: 4 ![]()
![]()
![]()
![]()
|
|
 |
|
A Danchin
Natural selection and immortality
Biogerontology (2009) 10: 503-516 ![]()
![]()
![]()
A Danchin
Bacteria as computers making computers
FEMS Microbiol Rev (2009) 33: 3-26 ![]()
![]()
![]()
A Danchin
Cells need safety valves
Bioessays (2009) 31: 769-773
![]()
![]()
In Escherichia coli, the role of lacA, the third gene of the lactose operon, has remained an enigma. I suggest that its role is the consequence of the need for cells to have safety valves that protect them from the osmotic effect created by their permeases. Safety valves allow them to cope with the buildup of osmotic pressure under accidental transient conditions. Multidrug resistance (MDR) efflux, thus named because of our anthropocentrism, is ubiquitous. Yet, the formation of simple leaks would result in futile influx/efflux cycles. Versatile modification enzymes with low sensitivity solve the problem if the modified metabolite is the one exported by MDR permeases. This may account for the pervasive presence of acetyltransferases, such as LacA, associated to acetyl-metabolite exporters. This scenario of constraints imposed by efficient influx of metabolites provides us with a model that should be followed when constructing synthetic cells
A Danchin, A Sekowska
Physico-chemical prerequisites for the construction of a synthetic cell
in: Synthetic Chemistry, May 26th - 30th, 2008, in Bozen, Italy
Beilstein Institut for the Advancement of Chemical Sciences (2009) 1-13
![]()
F Fourmentin, A Danchin
A sequel to "Cells need safety valves"
Symplectic Biology (2010) 1: 1
G Postic, A Danchin, U Mechold
Characterization of NrnA homologs from Mycobacterium tuberculosis
and Mycoplasma pneumoniae
Symplectic Biology (2010) 1: c390t9c12uxx.1 ![]()
speA, speB,
speE: A Sekowska, P Bertin, A
Danchin
Characterization of polyamine synthesis pathway in Bacillus subtilis
168
Mol Microbiol (1998) 29: 851-858 ![]()
speB,
argI: A Sekowska, A Danchin, JL
Risler
Phylogeny of related functions: the case of polyamine biosynthetic
enzymes
Microbiology (2000) 146: 1815-1828 ![]()
|
|
A Sekowska, HF Kung, A Danchin |
| Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction | |
| J Mol Microbiol Biotechnol (2000) 2:
145-177 |
EPC Rocha, A
Sekowska, A Danchin
Sulphur islands in the Escherichia coli genome: markers of the
cell's architecture?
FEBS Lett (2000) 476: 8-11 ![]()
JY Coppée, S
Auger, E Turlin, A Sekowska, JP Le Caer, V Labas, V Vagner, A Danchin,
I Martin-Verstraete
Sulfur-limitation-regulated proteins in Bacillus subtilis: a
two-dimensional gel electrophoresis study
Microbiology (2001) 147: 1631-1640
|
A Danchin
A path from predation to mutualism
Molecular Microbiology (2010) 77: 1346-1350 ![]()
|
|
ascR, snaA, tcyJ, tcyK, tcyL, tcyM,
tcyN, cmoO, cmoI, cmoJ, rbfK, sndA, ytnM :
CM Chan, A Danchin, P Marlière, A Sekowska Paralogous metabolism: S-alkyl-cysteine degradation in Bacillus subtilis Environ Microbiol (2014) 16: 101-117 |
|
|
A Danchin, A Sekowska The logic of metabolism and its fuzzy consequences Environ Microbiol (2014) 16: 19-28 |
PM Binder, A Danchin
Life's demons: information and order in biology. What subcellular
machines gather and process the information necessary to sustain life?
EMBO Reports (2011) 12: 495-499
In his seventeenth-century classic, Novum Organum, Francis Bacon wrote, “we cannot command nature except by obeying her” (Bacon, 2010). Although our knowledge of living systems is much improved since Bacon’s time, we are still far from understanding—or commanding—all the complex mechanisms of life. To take full advantage of living organisms for the benefit of mankind, we will need to understand those mechanisms to the furthest possible extent. To do so will require that the concept of information and the theories of information science take a more-prominent role in the understanding of living systems...
A Danchin
Les gènes du démon de Maxwell :
est-il possible de construire une usine cellulaire ?
Maxwell’s demon’s genes: Can we
build a cell factory?
Comptes Rendus de l'Académie des Sciences, Chimie (2011) 14:
413-419 ![]()
| A Danchin, A Sekowska The role of information in evolutionary genomics of bacteria In: Evolutionary genomics and systems biology (G Caetano-Anollés, Editor), Wiley & sons, Hoboken, NJ (2010) pp 81-94 |
A Danchin, PM Binder, S Noria
Antifragility and tinkering in biology (and in business): Flexibility
provides an efficient epigenetic way to manage risk
Genes (2011), 2:
998-1016; doi:10.3390/genes2040998 ![]()
M Porcar, A Danchin, V de Lorenzo, VA dos Santos,
N Krasnogor, S Rasmussen, A Moya
The ten grand challenges of synthetic life
Systems and Synthetic Biology (2011) 5:
1-9 ![]()
A Danchin
Scaling up synthetic biology: Do not forget the chassis
FEBS Letters (2012)586: 2129-2137. ![]()
A Danchin
Synthetic biology's flywheel
EMBO Reports (2012) 13: 92
![]() A
Danchin
A
Danchin
I geni del diavoletto di Maxwell
Prometeo (2012) 117: 120-125
|
|
CG Acevedo-Rocha, G Fang, M Schmidt, DW
Ussery, A Danchin From essential to persistent genes: a functional approach to constructing synthetic life Trends Genet. (2013) 29: 273-279. doi: 10.1016/j.tig.2012.11.001 |
|
|
|
V de Lorenzo, A Sekowska, A Danchin,
Chemical reactivity drives spatiotemporal organisation of bacterial metabolism FEMS Microbiology Reviews (2015) 39: 96-119 Abstract We examine how bacterial metabolism is shaped by chemical constraints acting on the material and dynamic layout of enzymatic networks and beyond. These are moulded not only for optimisation of given metabolic objectives (e.g. synthesis of a particular amino acid or nucleotide) but also for curbing the detrimental reactivity of chemical intermediates. Besides substrate channelling, toxicity is avoided by barriers to free diffusion (i.e. compartments) that separate otherwise incompatible reactions, along with ways for distinguishing damaging vs. harmless molecules. On the other hand, enzymes age and their operating lifetime must be tuned to upstream and downstream reactions. This time dependence of metabolic pathways creates time-linked information, learning and memory. These features suggest that the physical structure of existing biosystems, from operon assemblies to multicellular development may ultimately stem from the need to restrain chemical damage and limit the waste inherent to basic metabolic functions. This provides a new twist of our comprehension of fundamental biological processes in live systems as well as practical take-home lessons for the forward DNA-based engineering of novel biological objects |
|
|
|
G Boël, O Danot, V de Lorenzo, A Danchin Omnipresent Maxwell’s demons orchestrate information management in living cells Microb Biotechnol. (2019) 12: 210-242 doi: 10.1111/1751-7915.13378 |
|
|
|
V Courtier-Orgogozo, A Danchin, P-H Gouyon, C Boëte Preprint Evaluating the probability of CRISPR-based gene drive contaminating another species BioRxiv 776609 doi: 10.1101/776609. |
|
| P Lubrano, A Danchin, CG
Acevedo-Rocha From minimal to minimized genomes: Functional design of microbial cell factories pp. 177-210 In: Minimal Cells: Design, Construction, Biotechnological Applications (2019) edited by Alvaro R. Lara, G.Gosset Springer ISBN: 978-3-030-31897-0 |
||
|
|
Danchin A Isobiology: A variational principle for exploring synthetic life Chembiochem (2020) 21: 1781-1792 doi: 10.1002/cbic.202000060 |
|
|
|
Danchin A, Marlière P Cytosine drives evolution of SARS-CoV-2 Environ Microbiol (2020) 22: 1977-1985 doi: 10.1111/1462-2920.15025 |
|
|
|
Danchin A Three overlooked key functional classes for building up minimal synthetic cells Synthetic Biology (Oxford) (2021) 6: ysab010 doi: 10.1093/synbio/ysab010 |
|
|
|
Danchin A In vivo, in vitro and in silico: an open space for the development of microbe-based applications of synthetic biology Microb Biotechnol (2022) 15: 42-64 doi: 10.1111/1751-7915.13937 |
|
|
|
Danchin A, Huang JD SynBio 2.0, a new era for synthetic life: Neglected essential functions for resilience Environ Microbiol (2022) 25: doi: 10.1111/1462-2920.16140 |
| Back to top |
| Dernière mise à jour 5 Mars 2023 | Last update March 5th 2023 |