Freeman DYSON
Do not forget nitrogen, sulfur, phosphorus
Many different scenarios for the origin of life (abiogenesis) have been proposed over the years. Here I explore the specific features of an origin of life developing on mineral solids, based on the idea that early selective steps are essential, and that the metabolism originating on stones is ready to answer the puzzle of selection, as well as the need to make polymers (macromolecules).
Following the consistency of Freeman Dyson's argument in his book Origins of Life, I also show that metabolic reproduction must have preceded the replication of genetic material and propose that nucleotides were created by an escape from a nitrogen-fixing process. Furthermore, homochirality is a misleading problem, because maintaining a mixture of stereoisomers would cost much more information than spontaneous symmetry breaking. The chirality of amino acids is not a relevant issue: You should either drive on the left or the right side of a road, not both. The choice of which side of the road to drive on is often contingent: the reason continental Europe chose the right is simply because Napoleon decided it! Similarly, there are many constraints that limit life based on the chemistry of macromolecules. However, if a chiral set has been retained, it can impose chirality downstream. This is why the retention of l-amino acids has guided the selection of d-carbohydrates (as l-glycerate would count with l-serine in the translation process).
The atoms that can be used to this purpose are limited in number, and it is only sloppy experiments and media hype that let some to think that arsenic could be present in the backbone of nucleic acids. At best it could be included as a "decoration" of the standard phosphodiester backbone.
A further line of reasoning suggests that, as in the evolution of human artefacts, things tended to start big and awkward to progressively miniaturize. In The emergence of the first cells I propose a novel scenario, combining two types of RNA worlds (based in part on the work of Charles Kurland from Uppsala University):
Abstract The scenario proposed here builds on the critical need for compartmentalisation at the origin of life. In a first step, the surface of minerals compartmentalised and selected the reactive compounds that formed primitive metabolism. Subsequently, RNA molecules replaced mineral surfaces after the discovery of nitrogen fixation and the emergence of ribonucleotides, in parallel with a machinery for synthesis of peptides, coenzymes and lipids. Then, the RNA-metabolism world developed into an RNA-genome world based on RNA as informational templates rather than substrates. Bordered by lipids, the first cells were phagocytes, Protokarya, which put together two compartments stemming from the RNA-metabolism world (the cytoplasm) and the RNA-genome world (the nucleus). Emergence of stable deoxyribonucleotides allowed the clustering together of genes into chromosomes. Phagocytosis created the opportunity for an escape based on an alternative metabolism of membrane lipids and conquest of extreme environments, with the Archaea, and on the emergence of a robust and phagocyte-resistant envelope, with the Bacteria. Reductive evolution allowed bacteria with a modified enveloped to be phagocyted again as symbionts of Protokarya, leading to the final generation of the Eukarya. Continuation of horizontal transfer of the genetic material initially resulting from phagocytosis was carried on with the emergence of gene transfer via specialised conjugation machineries and viruses. DOI: 10.1002/3527600906.mcb.20130025
This was followed in 2017. by a more detailed view of the origin of the genetic coding process, where the idea that Protokarya were phagocytic organisms is further developed
Lecture given in Rio de Janeiro for the 100th
Anniversary of the death of Louis Pasteur (February 1995)
Presentation of the conference in
Rio (in French, autumn 1994)
![]() Slides
of the presentation (in English)
Slides
of the presentation (in English)
In Comparative genomics of extant bacterial genomes unravels a scenario of the origin of life (2007) we substantiate some of the hypotheses proposed here. In particular the idea that metabolism predated template-mediated replication of nucleic acids becomes an inevitable consequence of what we know of extant life. This also emphasised the importance of iron-sulphur clusters at the origin of central metabolic pathways that must have predated life as we know it today, and that are conserved in most living organisms, placing the sulfur atom at the very heart of life.
Louis Pasteur discovered that an original characteristic of organic matter was associated with life: organic molecules resulting from living processes were optically asymmetric. In contrast, artificially produced molecules were symmetrical. Life must therefore have involved a specific process to differentiate it from normal chemistry. Together with his own philosophical (and perhaps religious) convictions, this meant that it was hardly conceivable that life could arise from chemical matter, whether mineral or organic. Life had to come from life. Since it was well known that a broth left in the open gave rise to a variety of obviously living processes, this had to correspond to the pre-existence of living seeds that could multiply in the broth:
« J'ai la prétention de démontrer avec rigueur que dans toutes les expériences où l'on a cru reconnaître l'existence de générations spontanées, chez les êtres les plus inférieurs, où le débat se trouve aujourd'hui relégué, l'observateur a été victime d'illusions ou de causes d'erreur qu'il n'a pas aperçues ou qu'il n'a pas su éviter. » *
« I claim to demonstrate rigorously that in all the experiments in which the existence of spontaneous generations has been thought to be recognised, in the most inferior beings, to which the debate is now relegated, the observer has been the victim of illusions or causes of error which he has failed to perceive or to avoid. »
Because life is so sensitive to high temperature, it was easy to destroy the seeds in the broth, and, with appropriate technical constructions, to prevent reinfection of the broth by living seeds: if this hypothesis reflected truth, then a broth sterilised by heat would be stable in time, and would not lead to the de novo creation of life. In contrast, allowing the broth to be open to the air - where seeds were supposed to be present - would start the well-known multiplication process. This created a difficulty raised by Pouchet who showed that a boiled hay broth reaveled life after some time. This was in fact caused by heat-resistant spores of the bacterium Bacillus subtilis (named the "hay bacterium" in many languages). A further objection could be raised to this approach if the living principle was immaterial (i.e. had no weight permitting it to sediment into the broth): but this could be tested by using vessels open to this immaterial principle, where the opening could not permit a material seed to reach the broth (this was the origin of the famous Pasteur's "swan" shaped vials). This Pasteur carried out, and started both a new conceptual theoretical trend in the study of the origin of life, and an industrial process, known as " pasteurisation ".
What is the situation today, and can we predict the future trends of research in this domain of research? In spite of the demonstration by Pasteur that no life emerged from a broth, there is still a dominating model where a " prebiotic soup " is the prerequisite for life's birth. Many scientists have however stressed that life had rather to start from a mineral environment, had we to propose that it started on Earth. Among them, four major leaders should be considered: Desmond Bernal, who pointed out the importance of clays in mineral catalysis of organic matter [1]; Samuel Granick, who considered that photosynthesis had to be created on a solid surface, and use sulphur compounds as redox intermediates, and that we should analyze extant metabolism to extrapolate back to the origin [2]; Graham Cairns-Smith, who established clearly that a prebiotic soup would be poisoned by its very capacity to generate a large - much too large - variety of organic compounds, and who proposed the existence of a clay replicating material as predating our organic life [3]; and, more recently, Günter Wächtershäuser, who insisted on the fact that metabolism, at the surface of solid particles, should be seriously considered as the only possibility for generating life as we know it today [4].
I shall not here summarise the famous little book of Schrödinger [5], nor endeavour to define the laws of life, but place in the limelight four processes that must be intimately associated in all living entities. They are: metabolism, compartmentalisation, memory and manipulation [6]. The two former processes are organized by small molecules (a few tens of atoms at most), whereas the two latter are controlled by macromolecules (nucleic acids and proteins), associated via processes that are considered as transferring information. Thus, two spatial scales are intertwined in living processes, that develop at a mesoscopic scale.
It has been thought to be so difficult to reconcile all these processes together that a physicist like Freeman Dyson has even proposed that life originated twice [7]. This explains why most molecular biologists have simply forgotten to take into account metabolism and compartmentalisation as questions posed to all models of the origin of life, and have only considered proteins and nucleic acids. In recent years, the concept of "chassis" proposed by Synthetic Biology, is remarkably adapted to take into account both processes. At the conceptual level also, when comparisons have been made between life and Turing machines, the general principles for the construction of a self-replicating machine (the chassis) have nearly always overlooked the need for compartmentalisation and metabolism.
In contrast - and this should come out as a surprising conceptual defect in scientific thought - scientists get often very excited when one discovers yet another organic molecule in the cosmos, be it only an amino-acid, as if this gave us a clue for the origin of life! Following another trend the recent discovery of ribozymes [8] has been perceived as allowing us to solve the famous vicious circle, who is the first, the chicken or the egg, nucleic acids or proteins [9]? As a consequence life is now seen as having originated in an " RNA world " endowed of the metabolic functions that are displayed today by protein enzymes. In this context it is amusing to illustrate the scientific debate by two quotations expressing the most opposite views. In an argument similar to that of Cairns-Smith in his Genetic Takeover, Steven Benner for example wrote « arguments that attempt to extrapolate from modern biochemistry back to the origin of life are futile », [10], whereas Wächtershäuser described his own approach [11] as « a reconstruction of precursor pathways by retrodiction from extant pathways », as was proposed by Granick** in a scenario inspired by photosynthesis [2]. While the former hypothesis makes that it is more or less impossible to guess what happened at the origin, the latter can be used as a heuristic approach. Of course we do no know which is right, and it is likely that the present metabolism is both an archive and a palimpsest. A major question remains however, for all models involving an RNA world, that of the origin of nucleotides, and - this is not of minor importance either - that of the origin of membranes. This places us back to the question of metabolism, and to another chicken and egg paradox: which is the first, RNA or small precursors? Granick's and Wächtershäuser's models are meant to solve this issue, by placing metabolism of small molecules at the origin, using the selective power of solid surfaces, this, without requiring as Cairns-Smith did, the need for an ancestral mineral genetic replication process.
The important line of reasoning when thinking about life is to consider two main processes, reproduction of the cell machinery, with its compartments (what synthetic biologists name the chassis) and its metabolism, and replication of the genetic program. In his book Origins of Life, Dyson has convincingly shown that this means that in any scenario of origins, reproduction must have predated replication. In Synthetic Biology efforts most investigators are interested in the program, not in the machine. Yet efforts by some, such as Doron Lancet or Pier Luigi Luisi, aim at understanding the reproduction phase, by constructing mathematical and experimental models of what could have happened in the past.
In a nutshell, the model can be summarised as follows. Appropriate mineral surfaces, carrying an excess of positive charges, can select from an aqueous environment molecules that are negatively charged, mainly polycarboxylates and phosphates. These molecules are able to react together, and only those that are able to bind on the surface are kept for further chemical evolution. In this model, entropy-driven processes are important because, on a surface, they favour polymerisation, especially when it is caused by elimination of a water molecule (this is what usually happens in biological polymerisation) [4,11]. Here too, the model goes against the popular trend, the entropy rise being the positive element that creates the order necessary for life construction [12].
Extant metabolism allows one to substantiate the model by identifying clues for the first steps of a surface metabolism, stressing the importance of a few autocatalytic steps (because they provide a self-consistent means to stabilise the synthesis of those molecules that are further metabolised). In this model, coenzymes and nucleotides - molecules usually overlooked by scientists working on the origin of life - are of prime importance. The core of metabolism is made of triose-phosphates [4], and energy is derived from iron and sulphur redox transitions, leading to formation of a solid which has iron-sulphur clusters at its core, pyrite [11]. At this point it is necessary to investigate further the fate of solid particles: organic molecular species must have substituted for them. Cairns-Smith has proposed that RNA molecules, as polyelectrolytes that could mimic clays, are the obvious substitutes of surfaces [2]. Is it possible to find in present day RNA molecules, a class that could have played such a role? In 1975, Wong (now at the Hong Kong University of Science and Technology), describing the structure of a possible universal genetic code, proposed that transfer RNA molecules have played the role of a rigid holder allowing for local modification of substrates [13]. Since then, many examples of metabolic alterations involving transfer RNA have been discovered (or rediscovered).
I have proposed to name homeotopic transformation the in situ modification of non nucleotide residues carried by tRNA molecules. This accounts for the fact that different chemical groups can often be used to modify a given position of the molecule held by the tRNA molecule. Several examples illustrate this process: amidation of glutamic acid on tRNAgln, first described by Wilcox and Nirenberg [14], and also found in chloroplasts [15], and addition of hydrogen selenide on an activated tRNASecys, charged with activated serine for synthesis of proteins containing selenocysteine both in eukaryotes and prokaryotes [16-18]. Another well-known example of homeotopic transformation is the formylation of methionine carried by initiator tRNA in prokaryotes. This illustrates the involvement of intermediary metabolism in the control of macromolecular syntheses, as expected if metabolism is historically intimately associated with translation processes (see below) [19]. Finally, tRNA is also associated with many other metabolic processes that are not related to translation. For example it has been observed that charged lysine tRNA is involved in the synthesis of lipids [20], or that charged glutamic tRNA is necessary for synthesis of the heme precursor aminolevulinate in chloroplasts and in many bacteria [21-26].
Remarkably, charged tRNA molecules can also be required in reactions involving peptide bond formation in the absence of ribosomes. This is the case of synthesis of cell wall peptides in Staphylococci or Micrococci where tRNAser, tRNAthr or tRNAgly are involved [27-30]. N-modification of proteins by addition of leucine or phenylalanine residues has also been demonstrated in Escherichia coli [31,32]. Finally, degradation of ubiquitylated proteins requires, at least in some cases, the addition of arginine residues provided by charged tRNAarg [33-35].
![]() Note
added 15th october 2005: the
work of Dieter Söll is strongly substantiating this view.
Note
added 15th october 2005: the
work of Dieter Söll is strongly substantiating this view.
Note added 23d february 2007: analysis of the
core genome of Bacteria supports the present scenario for the
origin of life. A summary of this view has been presented at the
Institute for Systems Biology: ![]() Presentation (1.3 Mb)
Presentation (1.3 Mb)
Note added 7th july 2007: In depth analysis of genomes from Bacteria spanning the whole tree of life is consistent with the scenario described here. The genome splits into a paleome, that recapitulates the scenario of origin, and a cenome, that allows the cell to occupy a specific niche.
Note added 16th june 2009: The paleome has to be split into two parts. The part meant to replicate the genome and the part meant to reproduce the cell machinery and casings. The latter, in turn, is split into several groups of genes. Some manage chemical incompatibilities (chemical frustration) while the rest manages energy-dependent degradation of RNAs and proteins. The corresponding genes behave as coding for Maxwell's demons, chosing to protect what is young or functional.
Homeotopy in intermediary metabolism
Are there other, more general, traces of homeotopy in present day metabolism? If one follows the hypotheses of Granick [2], modified by Ycas [36], then more precisely stated by Jensen [37], that enzyme specificity evolved by recruiting proteins that already existed and catalyzed similar reactions, ancestral metabolic traits should be found in proteins that are grouped as similar in structure (and most probably in amino-acid sequence). It follows that in such families one could find traces of ancestral homeotopic processes.
As time elapses the number of such cases steadily increases, and their number is growing rapidly as genome programmes progress. Kaplan and Nichols, in 1983, discovered that synthesis of para-aminobenzoate and tryptophane was catalyzed by enzymes coded by genes trpD and pabA derived from a common ancestor [38]. Goncharoff and Nichols further developed this observation by showing that syntheses involving chorismate were performed by enzymes exhibiting a significant degree of similarity, such as enzymes synthesized from genes papB (para-aminobenzoate synthase) and trpE (anthranilate synthase) [39]. Further work by these authors and others showed that glutamine amidotransferase was involved in synthesis of para-aminobenzoate, tryptophane, as well as guanine (genes pabA, trpG and guaA) and derived from a common ancestor [40, 41]. A further substantiation of the existence of a primitive amidotransferase catalytic domain comes from analysis of human CTP synthetase, where the glutamine amide transfer domain is clearly related to the bacterial counterparts [42-44].
Subsequently Parsot, Cohen and their colleagues discovered that many activities involving pyridoxal phosphate were strongly related, in particular in synthesis or degradation of threonine, serine and tryptophane (thrC, dsdA, ilvA and trpB), as well as in enzymes involved in biosynthesis of methionine (metB, metC in E. coli, and their counterparts, in yeast) [45, 46]. In the same way Schoenlein et al. identified a significant level of similarity between enzymes responsible for the synthesis of pyridoxal phosphate (pdxB) and serine (serA) [47]. This is most revealing in view of Wächtershäuser's hypothesis of early surface metabolism, where triose-phosphates had to play a major role (and this contradicts Benner's dismissal of pyridoxamine as involved in early living processes [48]). Along the same line, we demonstrated that cysteine biosynthesis, in E. coli, shares a common ancestor with tryptophane biosynthesis [49]. This, together with the observation that cysteine and tryptophane codons are found in the same box of the genetic code table (in company with the UGA selenocysteine codon, also derived after homeotopic transformation from activated serine), substantiates the hypothesis that serine(-phosphate) was a general precursor of several amino-acids synthesis and that tRNA was involved in the process. Another old observation, the significant binding of charged tRNAleu or tRNAval to E. coli threonine deaminase, is well in line with this hypothesis [50]. Finally, we recently discovered that there is a significant kinship between synthesis of aspartyl-phosphate, glutamyl-phosphate, carbamyl-phosphate, and uridine-diphosphate in bacteria [51]. This is revealing when one remembers that aspartate and carbamyl phosphate are precursors of pyrimidines.
Peptides synthesising peptides
But all this does not tell us directly what could have been the precursors of these " holder " nucleic acids that seem to have played an early role in evolution of metabolism. It is clear that nucleotides are today part of several coenzymes, as pointed out by many [10, 52, 53]. But it cannot be surmised whether this reflects a trace of older structures rather than more recent adaptation of long nucleic acid precursors to shorter structures. Peptides are far much easier to synthesise than nucleic acids. On the other hand, many coenzymes (e.g. glutathione, pantothenate, folic acid ...) are (iso) peptides, or contain peptides, often at their active center. It is therefore interesting to explore whether (iso) peptides have not been precursors not only of most coenzymes, but of nucleotides as well. Many features of extant metabolism are pointing in this direction, for amino-acids are certainly present in the biosynthesis of purines and pteridines (glutamine, glycine, aspartate and serine, through formyltetrahydrofolate), or pyrimidines (aspartate).
But, as an indispensable self-catalytic step requires, are (iso) peptides involved in synthesis of peptides? The example of peptide antibiotics synthesis is a remarkable illustration of such self-referring catalysis. Biosynthesis of tyrocidin or gramicidin derives from formation of peptide bonds, in the absence of ribosomes. In tyrocidin, for instance, ten individual amino-acid residues are activated by ATP (as they are in translation) but are then transferred to an active SH residue of a protein subunit, forming a thioester bond. Tyrocidin synthesis begins after all ten sites of the enzyme complex have been esterified with their specific residue. The first three amino-acid residues react together sequentially, forming a thioesterified tripeptide. From this step onwards a phosphopantetheine cofactor transports the growing peptide chain on each new residue in turn, using its SH end as a carrier, and forms a new peptide bond following a transthiolation step, until the end of the process is reached when a decapeptide is formed (and finally cyclised). This process is highly reminiscent of synthesis of fatty acids from acetyl coenzyme A (which contains a phosphopantetheine arm as a reactive center). In this latter process acetyl-CoA is first transformed by carboxylation into malonyl-CoA using ATP as an energy source. A phosphopantetheine arm, bound to a core enzyme makes a succession of transthiolation reactions that lead to decarboxylation of malonate (this is the driving energy source) and condensation of two methylene residues on the growing chain. After six such steps the synthesis is completed, yielding palmitic acid. Thermal agitation supplies the only energy required for positioning of the carrier arm.
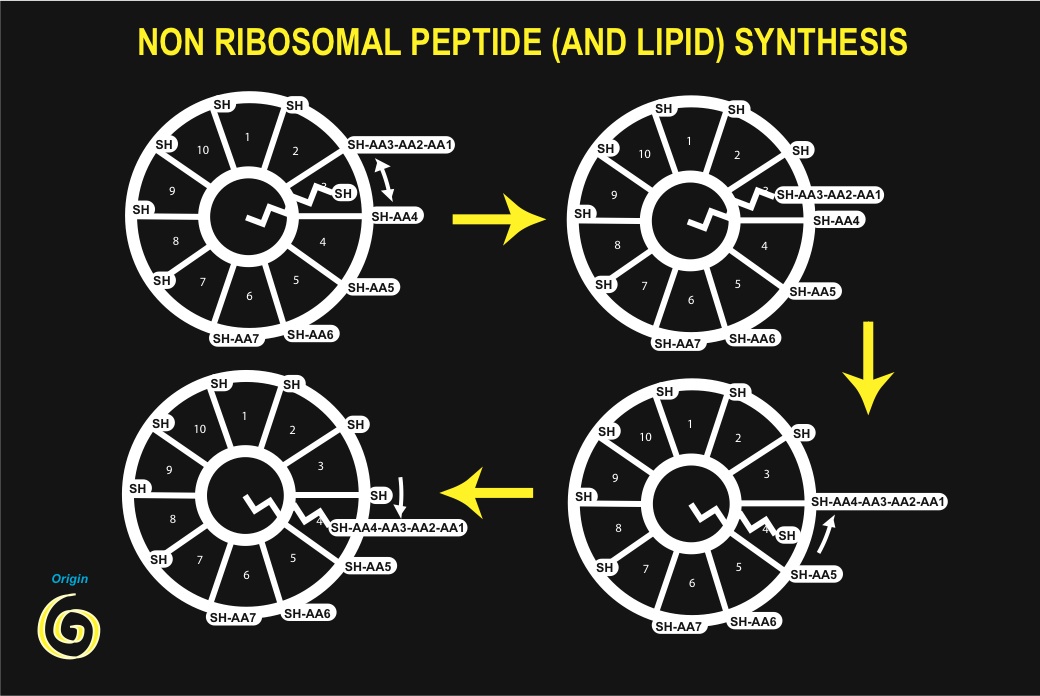
Analogy between both processes would only be anecdotal, had it not been discovered that indeed, proteins involved in tyrocidin, gramicidin and fatty acids synthesis share a common ancestor [54], indicating that their origin is common, and could be very old. This observation has been substantiated by the study of many other sequences, in particular from the programmes aiming at sequencing whole genomes [55]. Several features of these processes must be emphasized: (i) a peptide is able to carry out synthesis of a peptide; (ii) the same process permits synthesis of both lipids and peptides; (iii) the process requires the presence of active SH groups, essential components of surface metabolism as proposed by Granick and Wächtershäuser, and shown by De Duve to be of major importance for the origin of life [55]; (iv) energy is essentially derived from the formation of thioesters (and carboxylation / decarboxylation in the case of fatty acids); (v) among the amino-acids that are used, are present both L- and D-amino acids (as well as a basic amino-acid residue, ornithine, which cannot be incorporated into proteins during translation, because it cannot form stable adducts with tRNA).
If then, we accept that peptide formation is a very ancient process (it could predate the invasion of Earth by L-amino-acids), synthesis of peptide bond predating translation, it becomes particularly important to assess the hypothesis that nucleotides could have been derived from surface metabolites containing peptides. In this framework translation is a later invention, when tRNA molecules, instead of simply offering a general holding device for homeotopic transformation, have been involved in an RNA-mediated process for peptide-bond formation. A large number of examples where peptides can react by intramolecular reaction to form new molecules can be found. This is typical, for example of the antibiotic nisin or other lantibiotics, where serine and cysteine react to form lanthionin, a structural analogue of diaminopimelic acid [56-59]. Another example of this situation is the case of decarboxylases that use a pyruvoyl active site, derived from self-processing of a serine-serine dipeptide in the polypeptide proenzyme [60].
Nitrogen fixation and the origin of nucleotides
Organic molecules are usually presented as derivatives of carbon chemistry. Yet it is clear that the organic molecules present in living organisms display also a very high content of nitrogen. This was not thought to pose a difficult problem 40 years ago, when the models of the primitive atmosphere considered it to be strongly reducing, and rich in NH3. This is no longer the case. And one now considers that 3.8 billions years ago the atmosphere was mostly rich in CO2 and N2. It is therefore of the utmost importance for any model of the origin of life to propose scenarii permitting to understand prebiotic nitrogen metabolism.
Following Granick's approach, it may be interesting to analyze the
present day situation of nitrogen scavenging. In general, the
process requires the presence of iron-sulphur proteins such as
ferredoxins as electron transfer intermediates and molybdenum.
Ferredoxins are proteins constructed with a limited number of
amino-acid species, and they contain an iron sulphur cluster,
typical of what could be expected for very early proteins.
Molybdenum is a rare metal in today's earth crust, but it is not
clear whether at some stage of earth evolution, or locally it could
not have been abundant. In addition this metal ion could be replaced
by other ions for the electron transfer process at early stages of
nitrogen fixation [61]. The cofactor molybdopterin is found in
further complex oxidation reactions. Now, molybdopterin is a sulphur
containing coenzyme - that could be therefore be interacting with
metal-sulphur clusters, that is made of a pterin moiety derived from
GTP, by cyclisation following loss of a one-carbon formyl group.
This step is catalyzed by GTP cyclohydrolase, which yields pteridine
triphosphate from guanosine triphosphate with elimination of a one
carbon residue (precisely a residue transported by pteridine
containing coenzymes). Is it possible to consider that the reverse
reaction be a model for the straightforward synthesis of
nucleotides?
PteridineTP + HCOOH -----> GTP + H2O
We could then speculate that an autocatalytic process permitting synthesis of pteridine (tri) phosphate could have produced, as a side-product, the synthesis of GTP. In this frame of thought GTP would have but been a side-product of on-going nitrogen fixation.
This wild speculation would ask for a process permitting synthesis of pteridine from peptides. Exploration of extant microbial metabolism might give clues for the plausibility of such processes. The fact that many molybdopterin coenzymes contain nucleotides in their structure is already consistent with this hypothesis [62-64]....
Conclusion: a future for the origin of life
Several scientists convincingly proposed that life emerged from a surface metabolism, rather than from a poisonous broth. Granick's approach extensively used the knowledge of extant intermediary metabolism. If his main contention is right, this means that we are much nearer to the origin than we could think of previously. Analysis of biosynthetic pathways might therefore provide clues about the original metabolic pathways and processes. The hypothesis stating that early metabolism evolved through specification of broad range catalytic activities can be appreciated using comparison of enzyme structures in living cells. Among the most prominent processes are those which use tRNA molecules as carrier for homeotopic transformation of more or less universal precursors. Among such reactions, peptide bond formation could have evolved well before translation. Peptides are therefore placed in the limelight. They could have evolved before complex coenzymes, allowing their own synthesis. Synthesis of nucleotides would have been derived from nitrogen fixation (for purines and ribose) and from condensation of aspartate to carbamyl-phosphate (for pyrimidine synthesis). In turn nucleotides would have produced RNA carrier molecules, thus solving the chicken and egg paradox raised by the generally accepted hypothesis of an ancestral RNA world.
An interesting consequence of the depth of the conceptual reflection of scientists is that concepts can not only be organized to explain reality, but also to act in a creative way, constructing a new aspect of reality. This has been put into action for example when physicists have created Laser beams. Thus, nothing precludes that research in the Origin of Life results in new prospects: this is already illustrated by the success of selective combinatorial chemistry.
References
* Louis Pasteur (1862) "Sur les corpuscules organisés qui existent dans l'atmosphère. Examen de la doctrine des générations spontanées" in : Leçons de chimie et de physique professées en 1861 (à la Société chimique de Paris), Paris, Hachette et Cie, p. 219-254 (back to text)
1. Bernal JD (1951) The physical basis of life. Routledge and Kegan
Paul. London.
2. Granick S (1957) Speculations on the origin and evolution of
photosynthesis. Annals New York Acad. Sci. 69, 292-308.
November 2004:
In view of the fact that most investigators tend to think that traces
of the metabolic origin of life have been erased (in particular via
genetic takeover), it seems important to cite in extenso Granick's
view, which points out the fact that present metabolism is an archive
of the past, rather than a palimpsest, as this may be used as a
convenient heuristic approach to scenarios of the origin of life, as
demonstrated convincingly by Wächtershäuser in his model of surface
metabolism:
"I shall propose [...] that this unit
originated from some common minerals; that the minerals that
contain metal ions served both as coordinating templates and
catalysts for various reactions, and that around this unit were
formed organic molecules that gradually became organized into
units of ever-increasing complexity. Gradually, biosynthetic
chains developed in a stepwise fashion, using small molecules to
make molecules of ever-increasing complexity. The metal catalysts
became modified into the metalloenzymes; in these new complexes
the same metals would now act as more efficient catalyst.
The experimental method whereby it is proposed to find the
evolutionary precursors of protoplasm is to examine present-day
biochemical reactions in protoplasm and seek to relate them to
reactions that may have occurred and may still occur in the
minerals around us." ( back to text )
3. Cairns-Smith AG (1982) Genetic takeover and the mineral origin of
life. Cambridge University Press, Cambridge.
4. Wächtershäuser G (1988) Before enzymes and templates: theory of
surface metabolism. Mic. Rev. 52, 452-480
5. Schrödinger E (1944) What is life ? (reed. 1967) Cambridge
University Press, Cambridge.
6. Danchin A (1990) Une aurore de
pierres. Aux origines de la vie, Le Seuil, Paris.
7. Dyson FJ (1985) Origins of life. Cambridge University Press,
Cambridge.
8. Cech TR, Bass BL (1986) Biological catalysis by RNA Annu. Rev.
Biochem. 55, 599-637.
9. Danchin A (1983) L'Œuf et la Poule. Fayard, Paris.
10. Benner SA, Allemann RK, Ellington AD, Ge L, Glasfeld A, Leanz GF,
Krauch T, MacPherson LJ, Moroney S, Piccirilli JA, Weinhold E (1987)
Natural selection, protein engineering, and the last riboorganism :
rational model building in biochemistry. Cold Spring Harbor Symp.
Quant. Biol. 52, 53-63.
11. Wächtershäuser G (1990) Evolution of the first metabolic cycles.
Proc. Natl. Acad. Sci. USA 87, 200-204.
12. Danchin A (1986) Préface in Qu'est ce que la vie ? (translation of
E. Schrödinger What is life?) C. Bourgois, Paris.
13. Wong JTF (1975) A co-evolution theory of the genetic code. Proc.
Natl. Acad. Sci. USA 72, 1909-1912.
14. Wilcox M, Nirenberg M (1968) Transfer RNA as a cofactor coupling
amino-acid synthesis with that of protein. Proc. Natl. Acad. Sci. USA
61, 229-236.
15. Schön A, Gamini Kannangara C, Gough S, Söll D (1988) Protein
biosynthesis in organelles requires misaminoacylation of tRNA. Nature
331, 187-190.
16. Leinfelder W, Zehelein E, Mandrand-Berthelot MA, Böck A (1988)
Gene for a novel tRNA species that accepts L-serine and
cotranslationally inserts selenocysteine. Nature 331, 723-725.
17. Engelhardt H, Frochhammer K, Muller S, Goldie, KN, Böck A (1992)
Structure of selenocysteine synthase from Escherichia coli and
location of tRNA in the seryl-tRNA(sec)-enzyme complex. Mol.
Microbiol. 6, 3461-3467.
18. Baron C, Sturchler C, Wu XQ, Gross HJ, Krol A, Böck A (1994)
Eukaryotic selenocysteine inserting tRNA species support selenoprotein
synthesis in Escherichia coli. Nucl. Ac. Res. 22, 2228-2233.
19. Danchin A (1973) Does formylation of initiator tRNA act as a
regulatory signal in E. coli? FEBS Letters 34, 327-332.
20. Nesbitt III JA, Lennarz WJ (1968) Participation of aminoacyl
transfer ribonucleic acid in aminoacyl phosphatidylglycerol synthesis.
J. Biol. Chem. 243, 3088-3095.
21. Schön A, Krupp G, Gough S, Berry-Lowe S, Gamini Kannangara C, Söll
D (1986) The RNA required in the first step of chlorophyll
biosynthesis is a chloroplast glutamate tRNA Nature 324, 281-284.
22. Schneegurt MA, Beale SI (1988) Characterisation of the RNA
required for biosynthesis of delta-aminolevulinic acid from glutamate.
Plant Physiol. 86, 497-504.
23. Li JM, Brathwaite O, Cosloy SD, Russell CS (1989) 5-aminolevulinic
acid synthesis in Escherichia coli. J. Bacteriol. 171,
2547-2552.
24. Rieble S, Beale SI (1991) Purification of glutamyl-tRNA reductase
from Synechocystis sp. PCC 6803. J. Biol. Chem. 266,
9740-9745.
25. Kannangara CG, Andersen RV, Pontoppidan B, Willows R, von
Wettstein D (1994) Enzymic and mechanistic studies of the conversion
of glutamate to 5-aminolevulinate. Ciba Found. Symp. 180, 3-20;
discussion 21-25.
26. Hungerer C, Troup B, Romling U, Jahn D (1995) regulation of the hemA
gene during 5-aminolevulinic acid formation in Pseudomonas
aeruginosa. J. Bacteriol. 177, 1435-1443.
27. Roberts WSL, Strominger JL, Söll D (1968) Biosynthesis of the
peptidoglycan of bacterial cell walls VI. J. Biol. Chem. 243, 749-756.
28. Petit JF, Strominger JL, Söll D (1968) Biosynthesis of the
peptidoglycan of bacterial cell walls VII. J. Biol. Chem. 243,
757-767.
29. Roberts RJ (1974) Staphylococcal transfer ribonucleic acids. J.
Biol. Chem. 249, 4787-4796.
30. Green CJ, Vold BS (1993) Staphylococcus aureus has
clustered tRNA genes. J. Bacteriol. 175, 5091-5096.
31. Leibowitz MJ, Soffer RL (1971) Modification of a specific
ribosomal protein catalyzed by leucyl, phenylalanyl-tRNA: protein
transferase. Proc. Natl. Acad. Sci. USA 68, 1866-1869.
32. Shrader TE, Tobias JW, Varshavsky A (1993) The N-end rule in Escherichia
coli: cloning and analysis of the leucyl,
phenylalanyl-tRNA-protein transferase gene aat. J. Bacteriol.
175, 4364-4374.
33. Deutch CE (1984) Aminoacyl-tRNA: protein transferases. Methods
Enzymol. 106, 198-205.
34. Ferber S, Ciechanover A (1987) Role of arginine-tRNA in protein
degradation by the ubiquitin pathway. Nature 326, 808-811.
35. J. Li & C. M. Pickart (1995) Inactivation of arginyl-tRNA
protein transferase by a bifunctional arsenoxide: identification of
residues proximal to the arsenoxide site. Biochemistry 34, 139-147.
36. Ycas M (1974) On earlier states of the biochemical system. J.
Theor. Biol. 44, 145-160.
37. Jensen RA (1976) Enzyme recruitment in evolution of new function.
Annu. Rev. Microbiol. 30, 409-425.
38. Kaplan JB, Nichols BP (1983) Nucleotide sequence of Escherichia
coli pabA and its evolutionary relationship to trp(G)D.
J. Mol. Biol. 168, 451-468.
39. Goncharoff P, Nichols BP (1984) Nucleotide sequence of Escherichia
coli pabB indicates a common evolutionary origin of
p-aminobenzoate synthetase and anthranilate synthetase. J. Bacteriol.
159, 57-62
40. Kaplan JB, Merkel W, Nichols BP (1985) Evolution of glutamine
amidotransferase genes. J. Mol. Biol. 183, 327-340
41. Zalkin H, Argos P, Narayana SVL, Tiedeman AA, Smith JM (1985)
Identification of a trpG-related glutamine amide transfer domain in Escherichia
coli GMP synthetase. J. Biol. Chem. 260, 3350-3354.
42. Weng M, Makaroff CA, Zalkin H (1986) Nucleotide sequence of Escherichia
coli pyrG encoding CTP synthetase. J. Biol. Chem. 261,
5568-5574.
43. Mei B, Zalkin H (1989) A cysteine-histidine-aspartate catalytic
triad is involved in glutamine amide transfer function in PurF-type
glutamine amidotransferases. J. Biol. Chem. 264, 16613-16619.
44. Yamauchi M, Yamauchi N, Meuth M (1990) Molecular cloning of the
human CTP synthetase gene by functional complementation with purified
human metaphase chromosomes EMBO J. 9, 2095-2099
45. Parsot C (1986) Evolution of biosynthetic pathways : a common
ancestor for threonine synthase, threonine dehydratase, and D-serine
dehydratase EMBO J. 5, 3013-3019
46. Parsot C, Saint-Girons I, Cohen G (1987) Enzyme specialization
during the evolution of amino-acid biosynthetic pathways. Micro. Sci.
4, 258-262
47. Schoenlein PV, Roa BB, Winkler ME (1989) Divergent transcription
of pdxB and homology between the pdxB and serA
gene products in Escherichia coli K-12. J. Bacteriol. 171,
6084-6092
48. Benner SA, Ellington AD, Tauer A (1989) Modern metabolism as a
palimpsest of the RNA world. Proc. Natl. Acad. Sci. USA 86, 7054-7058
49. Lévy S, Danchin A (1988) Phylogeny of metabolic pathways:
O-acetylserine sulphydrylase A is homologous to the tryptophane
synthase beta subunit Molec. Mic. 2, 777-783
50. Singer PA, Levinthal M, Williams LS (1984) Synthesis of the
isoleucyl- and valyl-tRNA synthetases and the isoleucine-valine
biosynthetic enzymes in a threonine deaminase regulatory mutant of Escherichia
coli K-12. J. Mol. Biol. 175, 39-55
51. Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H,
Gilles AM. and Barzu O. (1995) Escherichia coli UMP-kinase, a
member of the aspartokinase family, is a hexamer regulated by guanine
nucleotides and UTP. Biochemistry 34, 5066-5074.
52. White HB, (1976) Coenzymes as fossils of an earlier metabolic
state. J. Mol. Evol. 7, 101-104.
53. Tremolières A (1980) Nucleotidic cofactors and the origin of the
genetic code. Biochimie 62, 493-496
54. Krätzschmar J, Krause M, Marahiel MM (1989) Gramicidin S
biosynthesis operon containing the structural genes grsA and grsB
has an open reading frame encoding a protein homologous to fatty acid
thioesterases. J. Bacteriol. 171, 5422-5429.
55. De Duve, C (1991) Blueprint for a cell. Portland Press, London.
56. Schnell N, Entian KD, Schneider U, Götz F, Zähner, Kellner R, Jung
G (1988) Prepeptide sequence of epidermin, a ribosomally synthesized
antibiotic with four sulphide-rings. Nature 333, 276-278.
57. Banerjee S, Hansen JN (1988) Structure and expression of a gene
encoding the precursor of subtilin, a small protein antibiotic. J.
Biol. Chem. 263, 9508-9514.
58. Buchman GW, Banerjee S, Hansen JN (1988) Structure, expression and
evolution of a gene encoding the precursor of nisin, a small protein
antibiotic. J. Biol. Chem. 263, 16260-16266.
59. Sahl HG (1994) Gene-encoded antibiotics made in bacteria. Ciba
Found Symp 186, 27-42.
60. van Poelje, PD, Snell EE (1990) Pyruvoyl-dependent enzymes. Annu.
Rev. Biochem. 59, 29-59.
61. Johnson JL, Rajagopalan KV, Mukund S, Adams MW (1993)
Identification of molybdopterin as the inorganic component of the
tungsten cofactor in four enzymes from hyperthermophilic Archaea. J.
Biol. Chem. 268, 4848-4852.
62. Karrasch, M, Borner, G, Thauer, RK (1990) The molybdenum cofactor
of formylmethanofuran dehydrogenase from Methanosarcina barkeri
is a molybdopterin guanine dinucleotide. FEBS Lett. 274, 48-52.
63. Johnson JL, Rajagopalan KV, Meyer O (1990) Isolation and
characterization of a second molybdopterin dinucleotide: molybdopterin
cytosine dinucleotide. Arch. Biochem. Biophys. 283, 542-545.
64. Borner G, Karrasch M, Thauer RK (1991) Molybdopterin adenine
dinucleotide and molybdopterin hypoxanthine dinucleotide in
formyltetrahydrofuran dehydrogenase from Methanobacterium
thermoautotrophicum (Marburg). FEBS Lett. 290, 31-34.