West African Studies
Mary H. KINGSLEY
![]()
A microbial metabolic pathways knowledge base
The European Union supports research through grants that allow the development of research activities involving several European partners, according to the principle of subsidiarity. The Microme program has been designed to support the construction of a platform for automatic and expert annotation of bacterial metabolic pathways. The effort presented here corresponds to our contribution via grant FP7-2009-222886-2.
This page aims to inform the general public (especially members of the European countries that run the European Union) about the latest developments of this research.
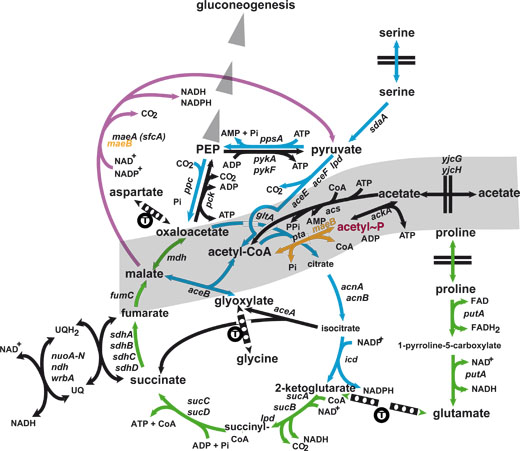
It has long been known that bacteria such as Escherichia coli, when grown on a carbon-rich diet in the presence of oxygen, respond in a biphasic manner, first growing exponentially, while accumulating acetate in the growth medium to compensate for the excess carbon, and then abruptly switching to an acetate-consuming phase when they enter the stationary phase. This process, called the « acetate switch », is the only known metabolic process that parallels consistently the entry into the stationary phase of growth. It is fairly universal, and corresponds to the function of the TCA « cycle » enzymes: during exponential growth, the pathway does not cycle, but produces metabolites essentially via the production of oxaloacetate and 2-ketoglutarate; then, in the respiratory phase, it cycles according to the classical TCA cycle presentation to produce protons allowing ATP synthesis and energy accumulation for the hard times to come (often in the form of polyphosphates).
After discussion with the geochemist Raoul-Marie Couture we wrote an article proposing a detailed scenario whereby some bacteria could synthesize monothioarsenate, a fairly innocuous derivative of arsenic. The metabolic scenario is hypothetical and fairly wild, of course, but showing that we should explore many biochemical hypotheses before trying to challenge our standard knowledge of the constraints of the law of physics on atoms. This paper is published in Environmental Microbiology. More about the arsenic nightmare.
Among the various metabolic functions we have to characterize for Microme are unknown, or unrecognized important functions. We have further identified nanoRNase families that are meant to supply for the degradation of the short leftovers of processive RNases. We also note that these enzymes interfere with the regulation of sulfur metabolism (via control of the reduction of sulfate), as well as lipid metabolism (via formation of 4-phosphopantetheine from Coenzyme A).
This is also an important knowledge for the construction of synthetic cells.
We note that this type of function deals with generic molecules (oligonucleotides) of a type that is not explicitly identified in metabolic databases, but that Microme aims at taking into account.
CM Chan, A Danchin, P Marlière, A Sekowska
Paralogous metabolism: S-alkyl-cysteine degradation in Bacillus
subtilis
Environ Microbiol (2014) 16: 101-117 doi:
10.1111/1462-2920.12210
A Danchin, A Sekowska
The logic of metabolism and its fuzzy consequences
Environ Microbiol (2014) 16: 19-28
doi: 10.1111/1462-2920.12270
CG Acevedo-Rocha, G Fang, M Schmidt, DW
Ussery, A Danchin
From essential to persistent genes: a functional approach to
constructing synthetic life
Trends Genet. (2013) 29: 273-279. doi:
10.1016/j.tig.2012.11.001
E Belda, A Sekowska, F Le Fèvre, A Morgat, D
Mornico, C Ouzounis, D Vallenet, C Médigue, A Danchin
An updated metabolic view of the Bacillus subtilis 168 genome
Microbiology (2013) 159: 757-770. doi:
10.1099/mic.0.064691-0
RM Couture , A Sekowska, G Fang, A Danchin
Linking selenium biogeochemistry to the sulfur-dependent biological
detoxification of arsenic
Environ Microbiol (2012) 14: 1612-1623. doi:
10.1111/j.1462-2920.2012.02758.x.
A Danchin
A path from predation to mutualism
Molecular Microbiology (2010) 77: 1346-1350
A Danchin
Scaling up synthetic biology: Do not forget the chassis
FEBS Letters (2012)586: 2129-2137.
doi: 10.1016/j.febslet.2011.12.024
A Danchin, PM Binder, S Noria
Antifragility and tinkering in biology (and in business): Flexibility
provides an efficient epigenetic way to manage risk
Genes (2011),
2: 998-1016; doi:10.3390/genes2040998
S Engelen, D Vallenet, C Médigue, A Danchin
Distinct co-evolution patterns of genes associated to DNA polymerase
III DnaE and PolC
BMC Genomics (2012) 13: 69.
doi: 10.1186/1471-2164-13-69
M Porcar, A Danchin, V de Lorenzo, VA dos
Santos, N Krasnogor, S Rasmussen, A Moya
The ten grand challenges of synthetic life
Systems and Synthetic Biology (2011) 5: 1-9
doi: 10.1007/s11693-011-9084-5
G Postic, A Danchin, U Mechold
Characterization of NrnA homologs from Mycobacterium tuberculosis
and Mycoplasma pneumoniae
RNA (2012) 18: 155-165 doi:
10.1261/rna.029132.111
H Rohde, J Qin, Y Cui, D Li, NJ Loman, M
Hentschke, W Chen, Fei Pu, Y Peng, J Li, F Xi, S Li, Y Li, Z Zhang,
X Yang, M Zhao, Peng Wang, Y Guan, Z Cen, X Zhao, M Christner, R
Kobbe, S Loos, J Oh, L Yang, A Danchin, GF Gao, Y Song, Y Li, H
Yang, J Wang, J Xu, the E. coli O104:H4 Genome Analysis
Crowd-sourcing consortium, MJ Pallen, J Wang, M Aepfelbacher, R Yang
Open-source genomics of an isolate from a german family outbreak of
Shiga-toxin-producing Escherichia coli O104:H4
New England Journal of Medicine (2011) 365: 718-724
27-28 February 2012
« Functional Analysis for Synthetic Biology. Metabolic Frustration is
Driving Compartmentalisation »
Frontiers
in Systems Biology II
Baptist University
The Croucher Foundation
1 - 2 December 2011
Hong Kong, SAR Hong Kong, China
« Ageing vs senescence: the fate of the cell factory »
Synthetic Biology (SB) puts together two separate entities, a program and a chassis. The majority of SB-related work deals with the program, not the chassis. We will deal with the chassis and ask the question: will scaling up synthetic processes be possible, and to what extent? Indeed, scaling up implies reproduction of the chassis, i.e. making similar copies. As in all systems this implies progressive ageing. Living cells have a knack to make that ageing differs from senescence. We will explore how rhis is possible, and why this has important consequences in terms of SB.
Working
Seminar in Conceptual Biology
Department of Mathematics
The University of Hong Kong
30 November 2011
Hong Kong, SAR Hong Kong , China
« Update on antifragility: a concept used in banking that may be
relevant to (synthetic) biology »
Bageco11
Bacterial Genetics and Ecology
Kerkyra, Greece
29 may - 2 June 2011
« Antifragility: a concept used in banking that may be relevant
to (synthetic) biology »
Shanghai, October 20th, 2010
« Natural selection and Maxwell's demon »
Basel, September 27th, 2010
« Metabolic and spatial frustration as a constraint for synthetic
biology »
Barcelona, September 6th, 2010
« Metabolic frustration as a constraint for synthetic biology »
Interfacing biology, technology and
society
Molecular traffic jams and the reproduction vs replication dilemma
25-27 August 2010
Copenhagen, Denmark
Gif sur Yvette, July 8th, 2010
A Danchin : « From EEC genes to Maxwell’s demon’s genes
»