A
new light of Alchymie: taken out of the fountain of Nature. To
which is added a Treatise of Sulphur
Micheel Sandivogius
The methionine salvage pathway
Fruit ripening is triggered by the production of a gas, ethylene. Studying ethylene production in apple fruit tissue, Baur and Yang noted that although methionine was in sufficient quantity for ethylene production for only a few hours, ethylene was produced for months, suggesting a methionine recycling pathway. In plants, this methionine salvage pathway has been known as the methylthioadenosine (MTA) or Yang Cycle at a time when little was known about the chemical steps in this pathway. Since no volatile sulfur was detected, it was assumed that the sulfur atom was recycled to replenish the methionine pool. It was later shown that, in fact, the methylthio- group of MTA was recycled to methionine via 5'-methylthioribose (MTR) [Murr 75 , Adams 77]. It was initially thought that the methylthio- group was transferred to a four carbon acceptor molecule such as homoserine to regenerate methionine. However, later studies showed that in plants, as in yeast cells, rat liver and bacterial extracts, the ribose moiety of MTA was also incorporated into the 2-aminobutyryl moiety of methionine.
Akiho Yokota and his colleagues, in Nara, Japan, unraveled the chemical intermediates in the pathway present in Bacillus subtilis. When in Hong Kong we developed the genetic analysis of the pathway in this organism, and later in Pseudomonas aeruginosa. In the final touch on the metabolism we established that the salvage cycle is irreversible, via formation and hydrolysis of 2-ketoglutaramate. We have recently (2018) written an update of the knowledge of the MSP.
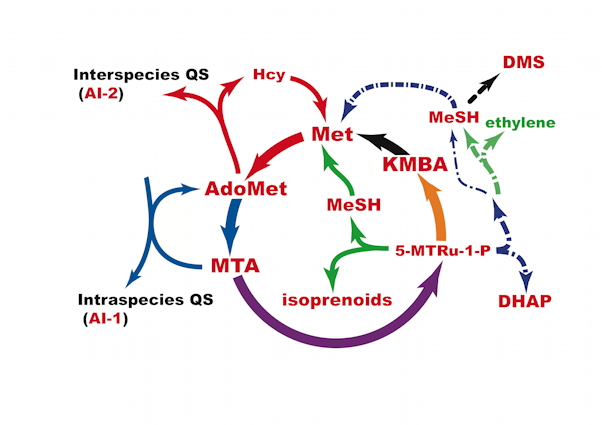
Methionine (Met) belongs to the twenty amino acids making proteins. It is also required for a great many cellular functions, including initiation of protein synthesis, methylation of DNA, rRNA, a variety of secondary metabolites and xenobiotics, and biosynthesis of cysteine (via the reverse transsulfuration pathway), phospholipids and polyamines. Polyamines, molecules with and aliphatic core and multiple amino groups, are synthesized in large amounts in cells, such as parasites, bacteria and cancer cells.
During polyamine synthesis of ethylene, polyamines and siderophores, methionine is consumed through the utilization of S-adenosylmethionine (AdoMet) which releases 5'-methylthioadenosine (MTA). Since the amount of methionine is typically limiting in cells and de novo synthesis of methionine is energetically expensive, it is important to be able to recycle this amino acid. Methionine regeneration from MTA plays an important role in sustaining the continued production of ethylene and polyamines.
In many organisms, the carbon atoms from the amino acid methionine skeleton do not come from the expected source, aspartate. The methionine salvage pathway is a ubiquitous biochemical pathway (it is present in humans) that maintains methionine levels in vivo by recycling the thiomethyl- moiety of methionine through a degradation pathway that leads from AdoMet through methylthioadenosine (MTA). The ribose ring of MTA provides the carbon skeleton for acyclic intermediates in the salvage pathway. Both of these enzymes were discovered in the laboratory of Abeles. The biochemistry of important enzymes of the pathway is developed in the laboratory of Yokota and his successors.
The methionine salvage pathway is universally used to regenerate methionine from 5’-methylthioadenosine (MTA), a by-product of some reactions involving S-adenosylmethionine. .
mtnN: A Sekowska, A Danchin
Identification of yrrU as the methylthioadenosine nucleosidase gene in Bacillus subtilis
DNA Res (1999) 6: 255-264
|
|
mtnK: A Sekowska, L Mulard, S Krogh, JK Tse, A Danchin |
| MtnK, methylthioribose kinase, is a starvation-induced protein in Bacillus subtilis | |
| BMC Microbiol (2001) 1:
15 |
|
|
mtnW, mtnX, mtnD, mtnE: A Sekowska, A Danchin |
| The methionine salvage pathway in Bacillus subtilis | |
| BMC Microbiol (2002) 2:
8 |
|
|
|
mtnW: H Ashida, A Danchin, A Yokota
Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism?
Res Microbiol (2005) 156: 611-618
|
|
mtnA, mtnB, mtnC, mtnP: A Sekowska, V Dénervaud, H Ashida, K Michoud, D Haas, A Yokota, A Danchin |
| Bacterial variations on the methionine salvage pathway | |
| BMC Microbiol (2004) 4:
9 |
mtnW: H Ashida, Y Saito, T Nakano, N Tandeau de Marsac, A Sekowska, A Danchin, A Yokota
RuBisCO-like proteins as the enolase enzyme in the methionine salvage pathway: functional and evolutionary relationships between RuBisCO-like proteins and photosynthetic RuBisCO
J Exp Bot (2008) 59: 1543-1554 doi: 10.1093/jxb/ern104
Finally, because methionine is costly to make, it is important to prevent its degradation. This result is achieved by the final step of the salvage pathway, where aminotransferase MtnV uses glutamine instead of glutamate as the alpha-amino group donor, producing alpha-ketoglutaramate. In turn, this metabolite is hydrolyzed into ammonium and alpha-glutarate that enters the TCA cycle, by omega-amidase (EC 3.5.1.3) MtnU. Because water is present at a 55 M concentration this step is, in practice, irreversible, preventing formation of the alpha-ketoacid precursor of methionine.
mtnU:
E Belda E, A Sekowska, F Le Fèvre, A Morgat, D Mornico, C Ouzounis,
D Vallenet, C Médigue, A Danchin
An updated metabolic view of the Bacillus subtilis 168 genome
Microbiology (2013) 159: 757-770. doi:
10.1099/mic.0.064691-0
A Danchin, A Sekowska
The logic of metabolism and its fuzzy consequences
Environ Microbiol (2014) 16: 19-28 doi:
10.1111/1462-2920.12270
A Sekowska, H Ashida, A Danchin
Revisiting the methionine salvage pathway and its paralogues
Microb Biotechnol. (2018) 12: 77-97 doi: 10.1111/1751-7915.13324