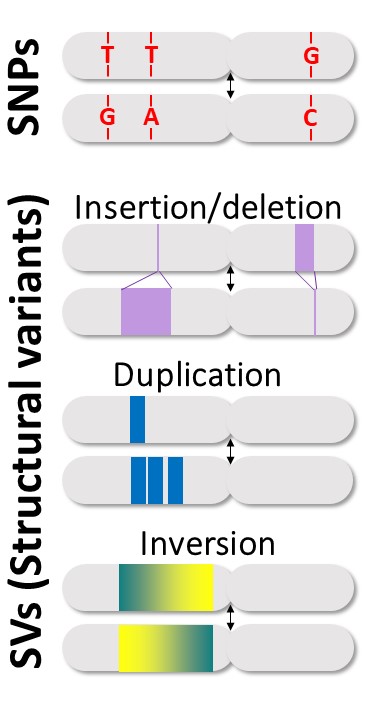
Genetic diversity is a fundamental level of biodiversity at a time of global change. It provides variation that underpins species persistence and their adaptation to changing environments. Variation in the direction or presence of DNA sequences has been largely overlooked until now. Yet, those structural variants (SVs) represent a key aspect of genetic diversity. SVs cover 3 to 10 times more of the genome than the well-studied single-nucleotide variants and have different properties (length, effect on recombination, mutation rate). Structural variation thus represents a quantitative and qualitative shift in our understanding of genetic diversity, with predicted, but understudied, implications for evolution.
The project EVOL-SV calls for a reassessment of the genomic basis of eco-evolutionary processes. We propose new research avenues on the impact of SVs in ecology and evolution and to determine the contribution of SVs to current biodiversity. My team combine cutting-edge genomics and powerful experimental approaches, developed throughout my career, to perform multidisciplinary research on a focal system, Coelopa flies, and then across taxa.
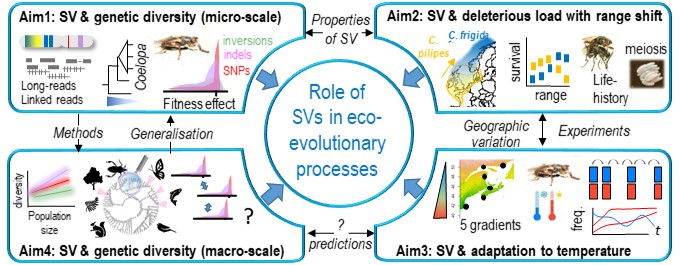
We investigate SVs within a population genetic framework in Coelopa spp. to determine how SV properties affect their distribution and effects on fitness. We assess the contribution of SVs to deleterious load in a case of range shift northwards. We examine how SVs contribute to phenotypic adaptation, focusing on parallel climatic gradients and rapid thermal variation. Finally, at a broader phylogenetic level, we will draw general principles about the evolution of structural genetic diversity.
We hope that EVOL-SV will have long-term impacts by providing the first comprehensive assessment of structural genetic diversity across the tree of life, developing the study of SVs in non-model species, and determining how genetic architecture contributes to evolutionary response in a rapidly changing world.
Species diversification and adaptation to heterogeneous environments have been suggested to be enhanced by chromosomal inversions, where a segment of the chromosome breaks, is reversed and then re-inserted. The most compelling aspects of inversions is reduced recombination in heterozygotes. As a result, inversions link together alleles and fonction as superloci, called supergene when they are associated with adaptive traits. They show intriguing patterns, such as varying in frequency along ecological clines, being involved in hybrid vigour, and harbouring an overabundance of loci involved in adaptation, population divergence and speciation.

An important axis of our research focuses on large polymorphic supergenes, formed by chromosomal inversions and maintained under balancing selection. We use a multi-layer integrative approach on the seaweed fly Coelopa frigida as a model species. By combining the population ecology, genomics and laboratory experiments, we investigate the association between inversions and adaptation to heterogeneous environments. we are also explicitely testing fitness in the lab and investigating functionally the genetic basis of local adaptation. Our aim is to shed light on the modalities by which very large chromosomal inversions contribute to adaptation in a non-model species and the selective forces and genetic mechanisms underlying the evolution of such structures.This project is undertaken by Léa Nicolas for her PhD. It is a thight collaboration with Pr. Pierre de Wit (University of Gothenburg, Sweden) and Dr Emma Berdan (Harvard, USA).
We are also interested in the evolution of this kind of supergene and chromosomal rearrangements at deeper phylogenetic level. In this collaboration with Dr Maren Wellenreuther (P&F research, New Zealand), we are thus studying Coelopidae at macro-evolutionary level to better retrace both genome and trait evolution in this group. This project is currently undertaken by Dominique Hicks for an internship.
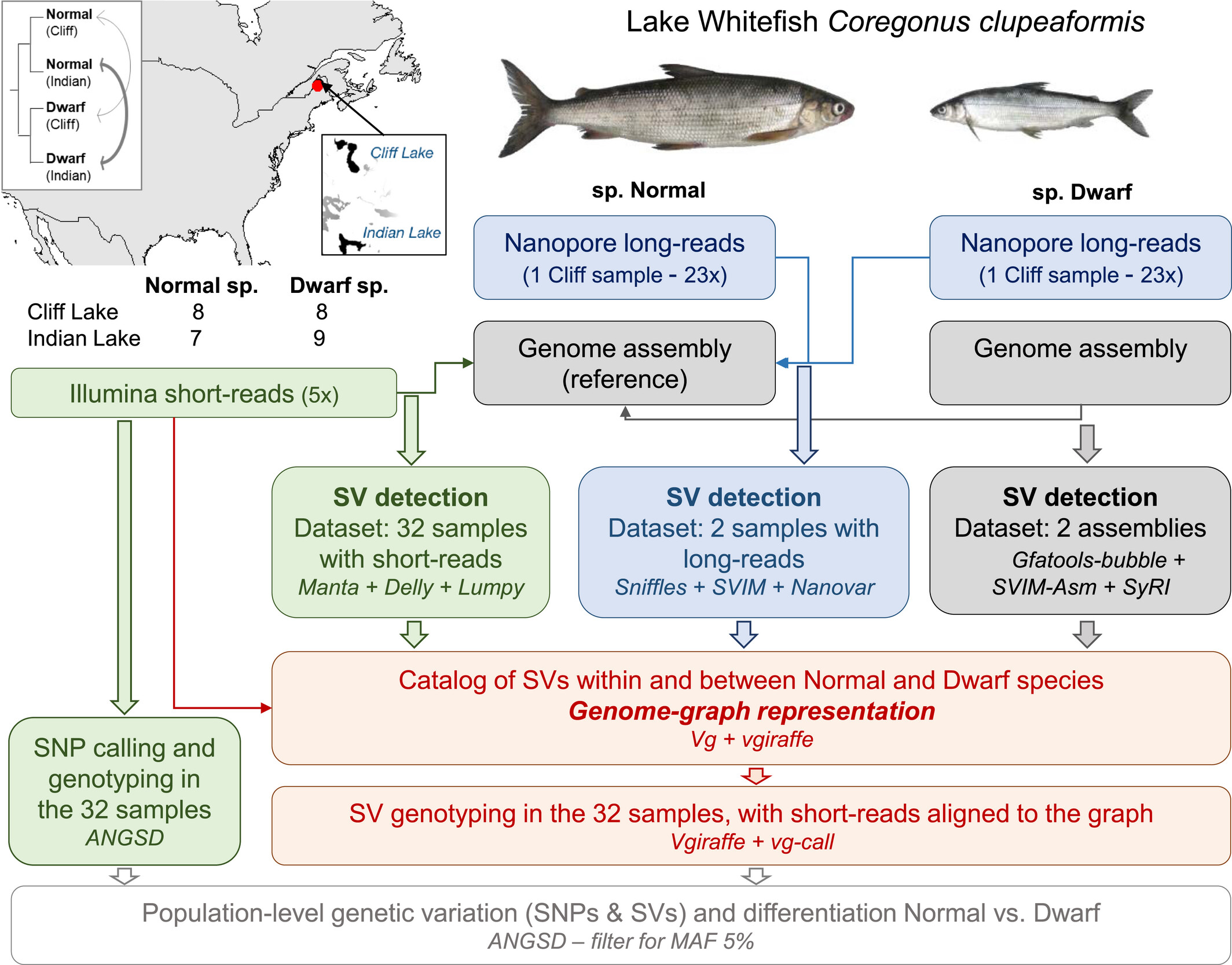
During my post-doc at University Laval with Pr. Louis Bernatchez, I have contributed to several studies on salmonids addressing the genetic architecture of adaptation or species differentiation with a particular focus on large chromosomal rearrangements and a range of structural variants.
For example, in the Lake whitefish Coregonus clupeaformis , we investigated the genetic of species differentiation and identified a preponderant role of transposable elements.
This study was also the opportunity to assess how different datasets (short-reads, long-reads and de novo assemblies) may help characterizing structural variants in the genome and to develop methods based on variant-aware genome graph to genotype SV in a population
My research focused on a big chromosomal inversion affecting multiple traits (sometimes called supergene) in the seaweed fly Coelopa frigida . By studying the population ecology, genomics and inversion frequencies in natural populations we investigated the implication of this inversion in local adaptation.
Then, using laboratory experiments, we explicitely tested fitness and modelled the different mechanisms of balancing selection that maintained the inversion polymorphism.
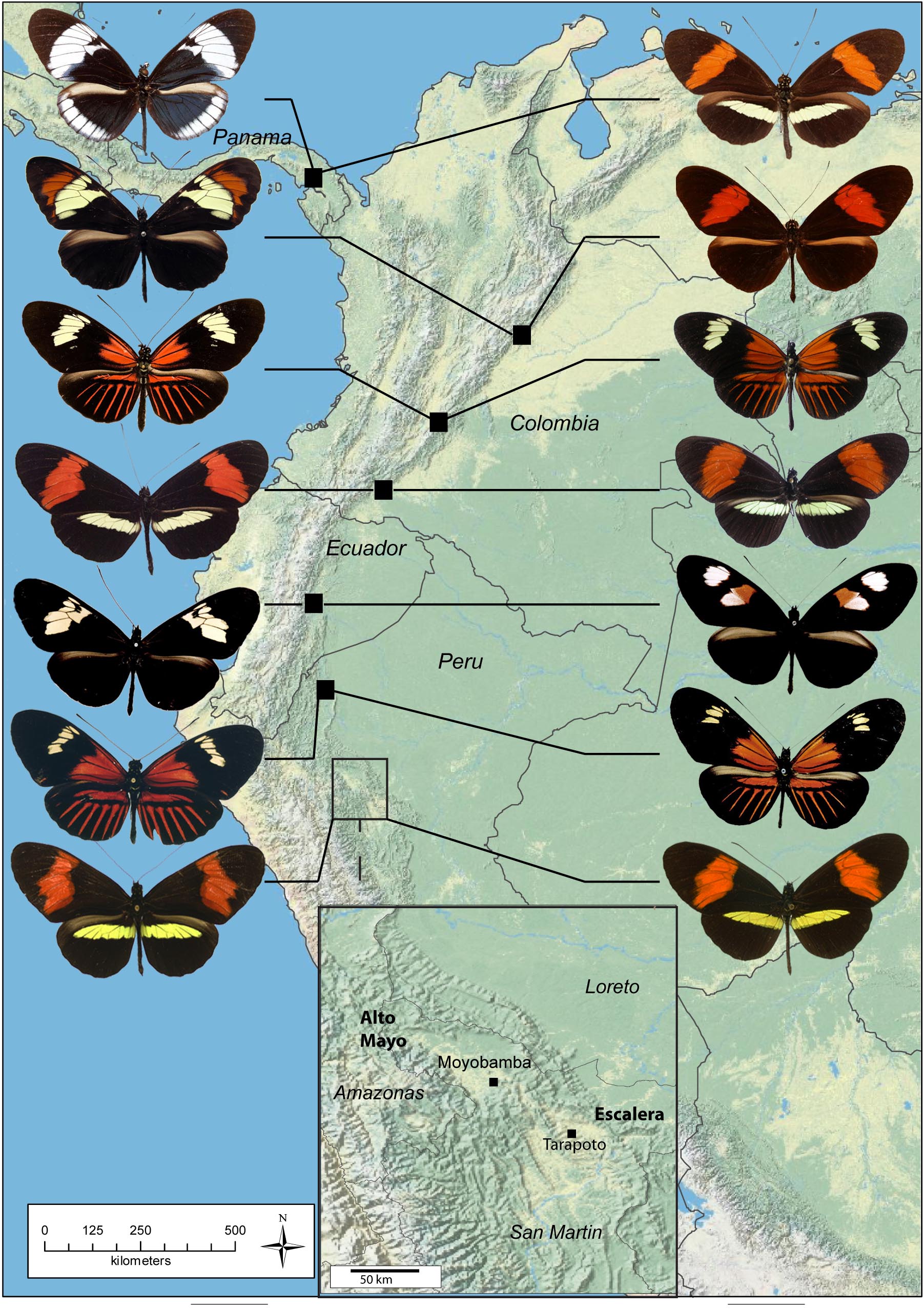
Recent radiations, such as the mimetic radiation of Heliconius butterflies in the Neotropics, offer an excellent example to address the evolutionary processes involved in speciation with gene flow. During my thesis, I explored an interesting situation in the Heliconius clade: two sibling species that are co-mimics of each other, sharing a similar wing colour pattern. This offers the possibility to study the limits of the classical model of speciation associated with shifts in wing pattern, usually described in this group. Colour pattern divergence indeed triggers strong reproductive isolation through disruptive natural selection for Müllerian mimicry and through sexual selection and assortative mating. I investigated how Müllerian mimicry affects species differentiation and reproductive isolation.
Specifically, I explored genetic divergence, coexistence, mimetic relationship and isolating barriers between the co-mimics H. timareta thelxinoe and H. melpomene amaryllis. I showed that the two species maintain consistent genetic and phenotypic divergence while hybridizing at low frequency. They display close resemblance in colour pattern, whose accuracy is affected by the composition of local mimetic community. Through controlled crosses, I showed that hybrids are intermediate in wing pattern and shape, suggesting that predation does not trigger strong post-mating isolation. However, behavioural sexual pre-mating isolation is high due to mate choice, likely relying on chemical cues.
Overall, my results confirmed that Müllerian mimicry between closely-related species likely enhances gene flow and acts against species differentiation. Nevertheless, this effect is balanced by the multidimensionality of isolating barriers, triggered by other ecological factors driving divergence.