Peter GODFREY-SMITH
H-NS
Microorganisms are able to thrive in environments differing by their temperature, osmolarity, pH, nutrient availability, etc. by rapidly adapting their structure and physiology. The corresponding processes are based on the existence of multiple regulatory networks that control gene expression in a coordinate manner in response to environmental stimuli, and in particular to changes in the environment.
Among the many genes that coordinate gene expression in gamma-proteobacteria the hns gene is particularly important. It has been characterized as a model system for studying the regulation of bacterial physiology. In Escherichia coli, the H-NS protein is involved in numerous cellular functions, affecting the expression of genes regulated by environmental factors or required in virulence.
Our experiments showed that proteins of the H-NS family are widespread in Gram-negative bacteria. Their structural and functional organization seems to be evolutionarily conserved. In particular, they are organized in two modules: the N-terminal part involved in oligomerization is specific for a bacterial family and linked to the C-terminal DNA-binding domain by a loop. At least in E. coli and related bacteria, H-NS-like proteins play a major role in bacterial physiology by controlling the expression of numerous genes such as those involved in bacterial motility. Most of them are regulated in response to environmental stresses, which suggests a role for H-NS in the maintenance of homeostasis, an essential condition for the survival inside their host.
![]() P
Lejeune, P Bertin, C Walon, K Willemot, C Colson, A Danchin
P
Lejeune, P Bertin, C Walon, K Willemot, C Colson, A Danchin
A locus involved in kanamycin, chloramphenicol and L-serine resistance
is located in the bglY-galU region of the Escherichia coli
K12 chromosome
Mol Gen Genet (1989) 218: 361-363
![]() P Bertin, P Lejeune, C Laurent-Winter, A Danchin
P Bertin, P Lejeune, C Laurent-Winter, A Danchin
Mutations in bglY, the structural gene for the DNA-binding
protein H1, affect expression of several Escherichia coli
genes
Biochimie (1990) 72: 889-891
![]() P Lejeune, A Danchin
P Lejeune, A Danchin
Mutations in the bglY gene increase the frequency of
spontaneous deletions in Escherichia coli K-12
Proc Natl Acad Sci U S A (1990) 87: 360-363
![]() P Bertin, P Lejeune, C Colson, A Danchin
P Bertin, P Lejeune, C Colson, A Danchin
Mutations in bglY, the structural gene for the DNA-binding
protein H1 of Escherichia coli, increase the expression of
the kanamycin resistance gene carried by plasmid pGR71
Mol Gen Genet (1992) 233: 184-192
![]() P Bertin, E Terao, EH Lee, P Lejeune, C
Colson, A Danchin, E Collatz
P Bertin, E Terao, EH Lee, P Lejeune, C
Colson, A Danchin, E Collatz
The H-NS protein is involved in the biogenesis of flagella in Escherichia
coli
J Bacteriol (1994) 176: 5537-5540
![]() JR Landgraf, M Levinthal, A Danchin
JR Landgraf, M Levinthal, A Danchin
The role of H-NS in one carbon metabolism
Biochimie (1994) 76: 1063-1070
![]() M Levinthal, P Lejeune, A Danchin
M Levinthal, P Lejeune, A Danchin
The H-NS protein modulates the activation of the ilvIH
operon of Escherichia coli K12 by Lrp, the leucine
regulatory protein
Mol Gen Genet (1994) 242: 736-743
![]() A Danchin, E Krin
A Danchin, E Krin
Filling the gap between hns and adhE in Escherichia
coli K12
Microbiology (1995) 141 ( Pt 4): 959-960
![]() C Laurent-Winter, P Lejeune, A Danchin
C Laurent-Winter, P Lejeune, A Danchin
The Escherichia coli DNA-binding protein H-NS is one of the
first proteins to be synthesized after a nutritional upshift
Res Microbiol (1995) 146: 5-16
![]() C Laurent-Winter, S Ngo, A Danchin, P Bertin
C Laurent-Winter, S Ngo, A Danchin, P Bertin
Role of Escherichia coli histone-like
nucleoid-structuring protein in bacterial metabolism and stress
response--identification of targets by two-dimensional
electrophoresis
Eur J Biochem (1997) 244: 767-773
![]() P Bertin, N Benhabiles, E Krin, C Laurent-Winter, C Tendeng, E
Turlin, A Thomas, A Danchin, R Brasseur
P Bertin, N Benhabiles, E Krin, C Laurent-Winter, C Tendeng, E
Turlin, A Thomas, A Danchin, R Brasseur
The structural and functional organization of H-NS-like
proteins is evolutionarily conserved in gram-negative bacteria
Mol Microbiol (1999) 31: 319-329
![]() O Soutourina, A Kolb, E Krin, C
Laurent-Winter, S Rimsky, A Danchin, P Bertin
O Soutourina, A Kolb, E Krin, C
Laurent-Winter, S Rimsky, A Danchin, P Bertin
Multiple control of flagellum biosynthesis in Escherichia
coli: role of H-NS protein and the cyclic AMP-catabolite
activator protein complex in transcription of the flhDC
master operon
J Bacteriol (1999) 181: 7500-7508 ![]()
![]() C Tendeng, C Badaut, E Krin, P Gounon, S Ngo,
A Danchin, S Rimsky, P Bertin
C Tendeng, C Badaut, E Krin, P Gounon, S Ngo,
A Danchin, S Rimsky, P Bertin
Isolation and characterization of vicH, encoding a new
pleiotropic regulator in Vibrio cholerae
J Bacteriol (2000) 182: 2026-2032 ![]()
![]() P Bertin, F Hommais, E Krin, O Soutourina, C
Tendeng, S Derzelle, A Danchin
P Bertin, F Hommais, E Krin, O Soutourina, C
Tendeng, S Derzelle, A Danchin
H-NS and H-NS-like proteins in Gram-negative bacteria and
their multiple role in the regulation of bacterial metabolism
Biochimie (2001) 83: 235-241
![]() F Hommais, E Krin, C Laurent-Winter, O Soutourina, A Malpertuy, JP
Le Caer, A Danchin, P Bertin
F Hommais, E Krin, C Laurent-Winter, O Soutourina, A Malpertuy, JP
Le Caer, A Danchin, P Bertin
Large-scale monitoring of pleiotropic regulation of gene expression
by the prokaryotic nucleoid-associated protein, H-NS
Mol Microbiol (2001) 40: 20-36
![]() F
Hommais, C Laurent-Winter, V Labas, E Krin, C Tendeng, O
Soutourina, A Danchin, P Bertin
F
Hommais, C Laurent-Winter, V Labas, E Krin, C Tendeng, O
Soutourina, A Danchin, P Bertin
Effect of mild acid pH on the functioning of bacterial
membranes in Vibrio cholerae
Proteomics (2002) 2: 571-579
![]() OA
Soutourina, E Krin, C Laurent-Winter, F Hommais, A Danchin, PN
Bertin
OA
Soutourina, E Krin, C Laurent-Winter, F Hommais, A Danchin, PN
Bertin
Regulation of bacterial motility in response to low pH in Escherichia
coli: the role of H-NS protein
Microbiology (2002) 148: 1543-1551
![]() C
Tendeng, OA Soutourina, A Danchin, PN Bertin
C
Tendeng, OA Soutourina, A Danchin, PN Bertin
MvaT proteins in Pseudomonas spp.: a novel class of
H-NS-like proteins
Microbiology (2003) 149: 3047-3050
![]() C
Tendeng, E Krin, OA Soutourina, A Marin, A Danchin, PN Bertin
C
Tendeng, E Krin, OA Soutourina, A Marin, A Danchin, PN Bertin
A Novel H-NS-like protein from an antarctic psychrophilic bacterium
reveals a crucial role for the N-terminal domain in thermal
stability
J Biol Chem (2003) 278: 18754-18760 ![]()
![]() F
Hommais, E Krin, JY Coppée, C Lacroix, E Yeramian, A Danchin, P
Bertin
F
Hommais, E Krin, JY Coppée, C Lacroix, E Yeramian, A Danchin, P
Bertin
GadE (YhiE): a novel activator involved in the response to
acid environment in Escherichia coli
Microbiology (2004) 150: 61-72
![]() C
Médigue, E Krin, G Pascal, V Barbe, A Bernsel, PN Bertin, F
Cheung, S Cruveiller, S D'Amico, A Duilio, G Fang, G Feller, C Ho,
S Mangenot, G Marino, J Nilsson, E Parrilli, EPC Rocha, Z Rouy, A
Sekowska, ML Tutino, D Vallenet, G von Heijne, A Danchin
C
Médigue, E Krin, G Pascal, V Barbe, A Bernsel, PN Bertin, F
Cheung, S Cruveiller, S D'Amico, A Duilio, G Fang, G Feller, C Ho,
S Mangenot, G Marino, J Nilsson, E Parrilli, EPC Rocha, Z Rouy, A
Sekowska, ML Tutino, D Vallenet, G von Heijne, A Danchin
Coping with cold: the genome of the versatile marine
Antarctica bacterium Pseudoalteromonas haloplanktis TAC125
Genome Res (2005) 15: 1325-1335 ![]()
![]()
![]()
![]() E
Krin, S Derzelle, K Bedard, M Adib-Conquy, E Turlin, P Lenormand,
M-F Hullo, I Bonne, N Chakroun, C Lacroix, A Danchin
E
Krin, S Derzelle, K Bedard, M Adib-Conquy, E Turlin, P Lenormand,
M-F Hullo, I Bonne, N Chakroun, C Lacroix, A Danchin
Regulatory role of UvrY in adaptation of Photorhabdus
luminescens growth inside the insect
Environmental Microbiology (2008) 10:
1118–1134
![]() Y
Jin, RM Watt, A Danchin, JD Huang
Y
Jin, RM Watt, A Danchin, JD Huang
Small noncoding RNA GcvB is a novel regulator of acid resistance in
Escherichia coli
BMC Genomics (2009) 10: 165
![]()
|
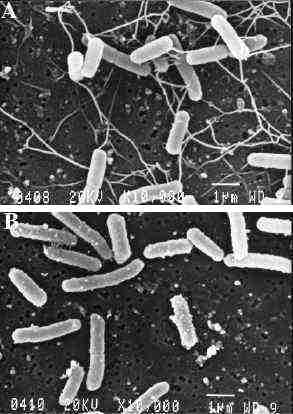
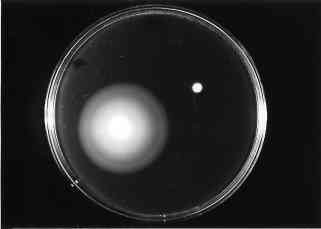
The swarming properties of E. coli hns strains are
altered on semi-solid medium (Figure
1 because of a lack of flagella (Figure
2. Their synthesis requires the expression numerous
genes organized in an ordered cascade. Their expression is
strongly reduced in a hns mutant. This suggests that
H-NS is required, directly or indirectly, in the expression
of flagellum biosynthesis genes (4).
Furthermore this is the first example showing that this
regulatory protein may have directly or indirectly a positive
control on gene expression.
|
||
 |
|
 |
|
Figure 1 : The presence of flagella visualized by scanning electron microscopy of wild-type (A) and hns (B) strains. (4) |
 |
Figure 2 : Motility test performed on E. coli wild-type and hns strains on semi-solid-medium. (4) |
|
The H-NS protein affects the expression of flhDC master operon which governs all flagellar genes. This mechanism is disctinct from the activation by the CAP/cAMP complex (8), ), requires the presence of an extended untranslated 5’ region (Figure 3 and is independent of StpA protein. (10). |
||
 |
||
| Figure
3: Regulatory region of flhDC master operon (8). The nucleotides are numbered relative to the transcriptional start site (+1). The CAP consensus sequence is indicated in red. Regions protected by H-NS are underlined by blue lines. The positions of the -10 and -35 boxes are indicated in yellow. The ATG translational initiation codon and the putative ribosome binding site (RBS) are indicated in green. |
||
| In V. cholerae, overexpression of the orthologous VicH protein strongly reduces motility. This, again, suggests a role for this H-NS-like protein in the control of motility. (9) | ||
|
In prokaryotes, the role of nucleoid-associated proteins
in bacterial physiology remains largely unknown. H-NS has
been initially isolated as a RNA polymerase associated
factor. It is a polyanion binding protein that may have
several regulatory targets
|
|
 |
|
|
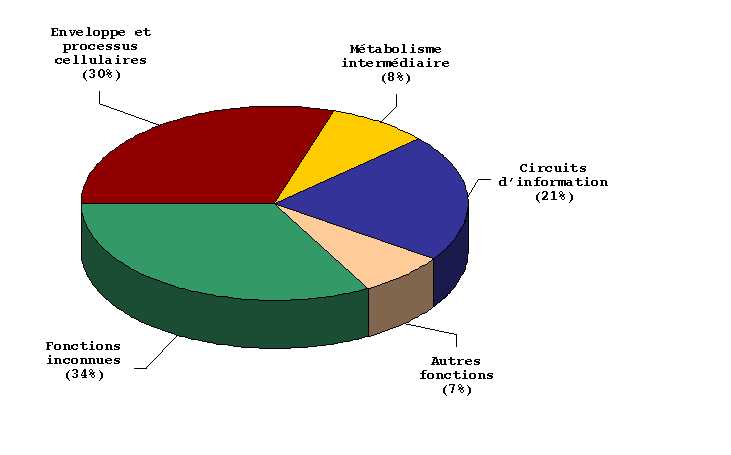
Figure 4:
Functional classification of H-NS-regulated genes.
|
|
|
By two-dimensional electrophoresis, the
synthesis and/or the accumulation of numerous proteins was
shown to be specifically affected by the hns
mutation (2).
Many proteins were identified by microsequencing procedure (5)
or, more recently, by mass spectrometry (11).
Most of them are involved in the adaptation of
microorganisms to environmental stresses such
as temperature, pH or osmolarity.
|
|
A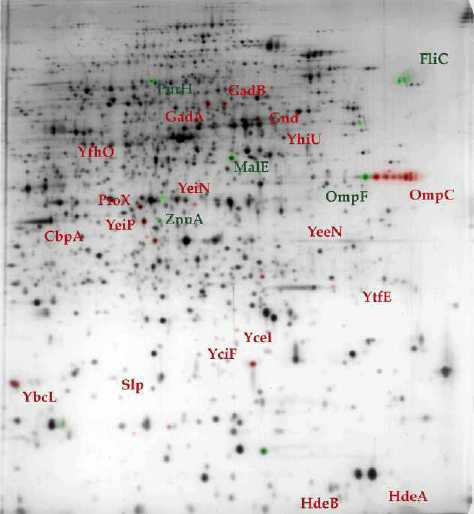 |
|
B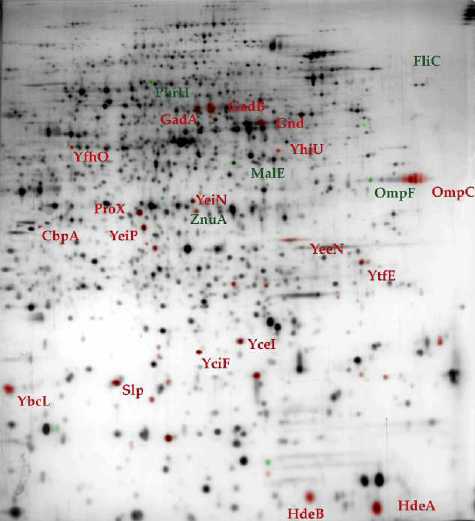 |
|
|
Figure 5 : 2D gel
electrophoresis of proteins extracted from wild-type (A)
and hns (B) E. coli strains . (11) In comparison with its wild-type counterpart, the accumulation level of proteins in green is reduced in hns strain while proteins in red are preferentially accumulated. |
|
|
The three-dimensional structure of H-NS protein has not
been yet determined, excepted for the C-terminal domain.
Nevertheless, in silico analysis suggests that this protein
contains 2 domains: the N-terminal part is predicted to be
mainly a-helical and to adopt a coiled-coil structure which
could play a role in oligomerisation and the C-terminal
DNA-binding domain which is predicted as a mixed a-b
structure. (Figure 6)
|
|
 |
|
|
A
|
|
|
B
|
|
|
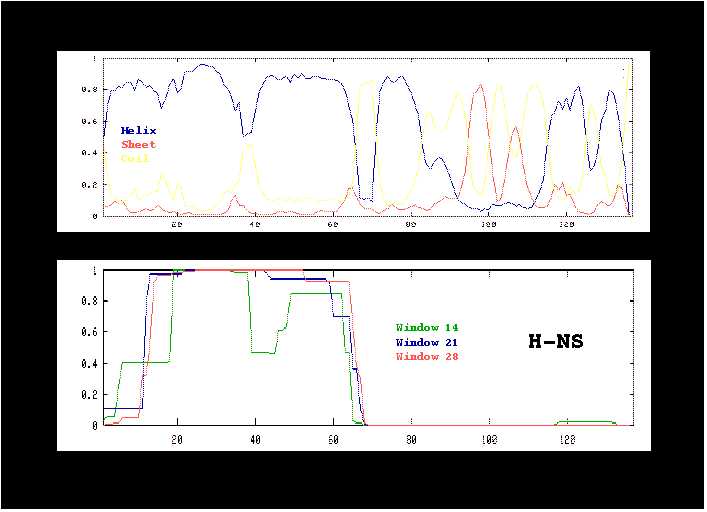
Figure 6:
Prediction of secondary structure by the MLRC method (A) and
of coiled-coil structure by the COILS program (B).
|
|
|
In silico analysis performed on
H-NS-like proteins of various microorganisms (6,7,9,10)
but also in vivo experiments (complementation of phenotypic
alterations associated with a hns mutation in E.
coli) suggest that they are structurally related (7).
The C-terminal part which is the most conserved region has been demonstrated to interact with DNA (Figure 7). All proteins show a preferential binding to curved DNA and, despite a low conservation in their N-terminal part, an ability to dimerize in vitro. These results demonstrate that a family of proteins structurally and functionally related to H-NS of E. coli exists in microorganisms, at least in Gram-negative bacteria (6,7,9,10). |
|
 |
|
|
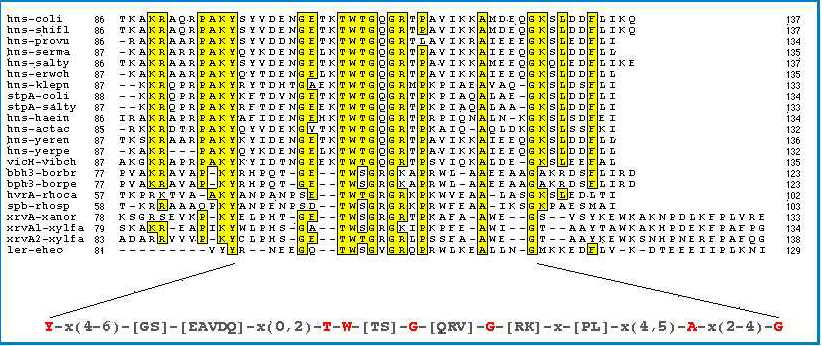
Figure 7:
Amino acid sequence alignment of the C-terminal domain of
H-NS and related proteins. Strictly conserved residues are
indicated in red. (10)
|
|