Teach Yourself
Genomics
•
Note that this page is under
permanent construction (2000-2003).
Genomics is a very fast changing field, and the landscape will be
entirely modified in a few years time.
 A
Brief History of the Human Genome Initiative
A
Brief History of the Human Genome Initiative
The history of the Human
Genome Initiative is deeply imbedded in sociology, psychology,
economy and politics. One can find several accounts of its course, and it
is probably too early to have a clear picture of the situation of the
revolution it is bringing about. The various
agencies involved in its setting up have their own historical
account.
Remarkably (and in a way that is generally not understood),
this programme probably started as a collaboration
between the USA and Japan, immediately after atomic bombs crushed
Hiroshima and Nagasaki, when questions were asked in the early fifties
about the future generation of people born from highly irradiated parents.
The Atomic Bomb Casualty commission, which was later included in the
activities of the Departement of Energy, initiated studies which played an
important role at the onset of the Human Genome Program. This question has
been recently asked again, and, of course, knowledge of the human genetic
polymorphism brought about by the sequencing of Single Nucleotide
Polymorphic (SNP) sites will boost again investigating the question. A
personal view of this programme, as a technical feat as opposed to a
concrete hypothesis-driven scientific programme can be found in The
Delphic Boat, what genomes tell us, Harvard University Press, 2003.
The term Genomics
appeared in the literature in 1988, but it was probably used well before
that time.
 Genome Sequencing
Genome Sequencing
A general description exists in French in La
Barque de Delphes, some is translated and adapted here. An
adaptation of this book is published at Harvard University Press, under
the title The Delphic Boat.
For a general strategy for sequencing bacterial genomes
see here.
DNA (standing for DeoxyriboNucleic Acid) is made of the
chaining together of four similar chemicals named nucleotides (or
abbreviated into the common but somewhat misleading expression "bases").
Four types of nucleotides, noted A, T, G and C form the DNA molecule.
The DNA molecule is made of two strands twisted around each other as in
an helical staircase:
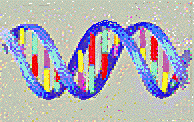
The chain makes a long thread, which can be described as
an oriented text, for example: >TAATTGCCGCTTAAAACTTCTTGACGGCAA>
etc. which faces the complementary strand
<ATTAACGGCGAATTTTGAAGAACTGCCGTT<
This is because the building blocks (which can be
referred to as "letters") are of four, and only four, types, and that A
always faces T and G faces C.
Genomic DNA is a long molecule, composed of hundreds of
thousands, millions or even billions of bases. Its chemical properties
are original (they are those of a long thread-like polymer), and this
allows DNA to be fairly easily isolated.
Isolating DNA
Cells are disrupted by physico-chemical means and DNA is
easily separated from the bulk because it is insoluble in alcohol or other
organic solvents. The difficult steps in isolating DNA is to break up the
cells, without altering the integrity of DNA (it is an extremely thin
molecule, the double helix cylinder having a diameter of about 2
nanometers, i.e. 2 millionth of a millimeter). In fact, DNA can
easily be seen as a thread-like molecule with the naked eye, and any type
of strong stirring or frequent pipetting will shear this fragile polymer.
Separation from proteins is achieved by adding enzymes (proteases, such as
those found in washing machine powders) which dissolve proteins into
water-soluble pieces. Further solvent extraction allows one to obtain DNA
precipitated in 70% alcohol. It is collected by centrifugation and
thoroughly washed with a water-alcohol mixture containing appropriate
chemicals meant to inactivate the agents which might degrade DNA. The
protocol is delicate, but it can be performed even in an hotel room (this
has happened!) with very simple equipment. DNA can be kept as such in an
ordinary refrigerator.
With the advent of fast sequencing as well as fast and heavy duty
computing facilities, it became possible to just fragment a genome in a
random fashion, to submit each fragment of a library to sequencing. This
is called "shotgun" sequencing.
The Polymerase Chain Reaction (PCR)
For an interesting history of the PCR, read the excellent
book by Paul Rabinow "Making PCR".
The usual physico-chemical systems we are familiar with
differ from living organisms, because the latter multiply by reproduction
of an original template. In nature, dispersed systems tend to be found at
lower and lower concentration from the original dispersion point as time
elapses. In contrast, a single living cell, that is allowed to grow in a
given environment, can multiply at a considerable level. In particular, as
soon as a DNA fragment is cloned into a replicative element, it can easily
yield 106, 109 or even 10^12 copies of itself. It is
this fundamental property which explains that one successfully determined
the sequence of many genomes. This is also the basis of the (not so
irrational) fear of the public for dispersion of genetically modified
organisms (GMOs). But although the techniques of in vivo molecular
cloning are indispensable to research in molecular genetics, they are
techniques tedious and difficult to put into practice. They are not always
possible to implement, and they are time consuming (one must isolate the
fragments to be cloned, incorporate them into replicative units, then
place these units into living cells that one must subsequently multiply).
Fortunately there exists, in particular for sequencing purposes, an
extremely efficient technique, Polymerase Chain Reaction (PCR), that
permits amplification in vitro and no longer in vivo of
any DNA fragment that can be obtained very rapidly in considerable
quantity.
With this technique, the fragment to be amplified is incubated in the
presence of a particular DNA polymerase which is not inactivated at high
temperature, together with the nucleotides (the basic DNA building blocks)
necessary to the polymerization reaction, and with short fragments
(oligonucleotides) complementary to each one of the extremities of the
region to amplify. The trick of the method is to use a polymerase that is
stable at high temperature (isolated from bacteria growing at very high
temperature, up to 100°C and more), and to repeatedly alternate cycles of
heating and cooling of the mixture. Typically, when the DNA double helix
is heated up, its two strands separate. One then cools down the mixture in
the presence of a large excess of oligonucleotides (synthesized chemically
and present in large excess in the assay mixture) that will each hybridize
to the sequence they must recognize at the extremity of the strand to
amplify. In the presence of the polymerase and of nucleotides, these
oligonucleotides behave as primers for polymerization (at the chosen
temperature sufficiently low to allow for hybridization). One subsequently
heats up the mixture to separate the newly polymerized chains, then cools
it down. The newly synthesized strands can therefore become new templates,
and the cycle repeats itself.
One thus understands that, in principle, one doubles at each cycle the
quantity of fragment obtained. At this rate, ten cycles would amplify the
fragment one thousand fold (210 times). Forty cycles or so give
therefore a very significant quantity of the fragment (there is not an
exact doubling at each cycle, of course, and the value obtained, although
very high and visible on a molecular size separation gel, will be somewhat
lower than that predicted by a doubling at each cycle, 240: one
thousand billion times). This method is extremely powerful since it can
amplify very low quantities of DNA (theoretically only one starting
molecule is enough). Since its invention in 1983, until the beginning of
the nineties, PCR could only yield short fragments (a few hundred base
pairs), and it displayed a high error rate (of the order of one wrong base
per one hundred), but the use of new DNA polymerases, together with better
tuned experimental conditions have considerably improved its performance.
Mid-nineties it was thus possible to obtain with polymerization chain
reaction, amplification of up to 50 kb-long segments, with a very low
error rate (one in ten thousand base pairs). It remained however to be
carefully verified that these fragments did not contain short deletetions,
following an accidental "jump" of the polymerase on its template.
Thus, PCR is a rapid and cheap technique very easy to put into practice,
that permits replication of a DNA fragment in a test tube, one million
times or more. Because of its simplicity PCR is important, for in
particular it can be performed by practically anybody, and amplify without
previous purification a DNA target present in very low quantity in almost
any medium, a billion times in a few hours. This technique had therefore a
huge impact in domains as varied as identification of pathogens, study of
fossil species disappeared for millions of years, genetic diagnosis or
crime detection (forensic investigations). For our problem, the sequencing
of genomes, PCR permits amplification of genome sequences in regions that
are impossible to amplify by cloning. In contrast to DNA obtained by PCR,
independent of life, DNA clones that multiply in cell lineages may, as we
observed in authentic situations, result in toxic properties. For example
the DNA of the bacteria that are used to make natto (fermented soy beans)
in Japan, Bacillus subtilis, is very toxic when it is cloned in a
vector replicating in the universal host for sequencing programs, Escherichia
coli. The underlying biological reason is simple, as it has been
discovered during the sequencing of the B. subtilis genome: the
signals that indicate that gene can be expressed into proteins (the
transcription and translation starts in B. subtilis) are
recognized in such a strong and efficient way in E. coli, that
they override the essential syntheses of their host in profit to those of
the segments cloned in the vector, leading the host to its premature
death.
Vectors
Sequencing DNA supposes amplification of the fragments
which will be replicated by DNA polymerase in the presence of appropriate
nucleotide markers (radioactive or fluorescent). Amplification can be
performed in vitro, using PCR, but the most common way to amplify
a DNA fragment is to place it into an autonomous replicating unit, which,
placed in appropriate cells, is amplified as cell multiplies while it is
carried over and copied along the generations, and found in the progeny as
a colony, a clone (hence the word "cloning" to illustrate the
process).
Making libraries
Sequencing
It is the possibility to perform in vitro what is
usually going one in vivo that gave to Frederick Sanger, from the
Medical Research Council in Cambridge in England, the idea that one could
alter the process in such a way as to determine the succession of the
bases of a template DNA. Sanger's simple, and for this reason bright, idea
is that it would be enough to start the reaction always at the same place,
and then to stop it precisely but at random times after each base type, in
a known and controlled fashion, to obtain in a test tube a mixture
comprising a large quantity of sequence fragments, each starting at the
same position in the newly polymerized strand, but ending randomly after
each of the four base types. For example, let us consider the following
sequence:
GGCATTGTTAGGCAAGCCCTTTGAGGCT>
If one knew how to stop the reaction specifically and randomly, after each
A one would get in the test tube the following fragments:
GGCA>
GGCATTGTTA>
GGCATTGTTAGGCA>
GGCATTGTTAGGCAA>
GGCATTGTTAGGCAAGCCCTTTGA>
In the same way, after each C:
GGC>
GGCATTGTTAGGC>
GGCATTGTTAGGCAAGC>
GGCATTGTTAGGCAAGCC>
GGCATTGTTAGGCAAGCCC>
GGCATTGTTAGGCAAGCCCTTTGAGGC>
and so on. And if one knew how to screen these fragments simply using
their mass (their length), then to place them in a row according to their
increasing or decreasing length, then one could construct four different
scales corresponding to each type of base at which each step would stop
specifically a after a given base type (A, T, C, or G). Placed side by
side (if the screen of each base type specific reaction was performed in
parallel experiments) they would thus provide directly the studied
sequence (telling that the fourth base is an A, the third a C, etc), and
this without asking the screening process to know the nature of the
considered base but only the length of the fragments that contain it.
Frederick Sanger is an excellent chemist, and he thought of a simple idea
to construct in practice this mixture of fragments, interrupted precisely
after each of the base types. As a start point, he synthesized chemically
a short strand, complementary to the beginning of the strand that he would
like to sequence. This is the primer, which will be in common with all
newly synthesized strands, making them all start at exactly the same
position. Then, to stop the polymerization reaction after a given base
type, he used chemical analogs of each nucleotide, in such a way as they
are recognized by DNA polymerase in a normal and specific way (they base
pair with a base in the complementary strand with the normal pairing
rule). These analogs are chosen so that they are incorporated in the
strand in the course of normal DNA synthesis, but their structure is such
that they prevent polymerization immediately after they have been
incorporated into a nascent polymer. Finally, to obtain a statistical
(random) mixture of fragments synthesized in this way (one must obtain all
possible fragments interrupted after each base type), he mixed up the
normal nucleotide (which, of course, does not stop synthesis after it has
been incorporated in the growing polymer) with the chain terminator
nucleotide, in proportions carefully chosen to let the reaction proceed
for some time before being stopped, and give a mixture of all types of
fragments, containing long fragments as well as short fragments. This
method is named after the chemical nature of the analogs ("dideoxy
nucleotides"): Sanger method, chain termination method or DNA polymerase
dideoxy chain terminator method.
Thus, by using an analog of A (here noted A*), one obtains, for example, a
mixture of the fragments shown above, with sequence GGCATT as a primer,
and wishing to copy the following strand:
>AGCCTCAAAGGGCTTGCCTAACAATGCC>
(which should be written
<CCGTAACAATCCGTTCGGGAAACTCCGA<
when read from right to left, to make clear the complementarity rule
between the bases of each strand):
GGCATTGTTA*>
GGCATTGTTAGGCA*>
GGCATTGTTAGGCAA*>
GGCATTGTTAGGCAAGCCCTTTGA*>
And the experiment will consist in performing in a concomitant way the
same reaction with the other analogs, G*, C* and T*.
The subsequent step consists in separating the different fragments in the
mixture, and to identify them. Separation is easily performed using
electrophoresis, by making the mixture migrate in a strong electric field,
through the mesh of a gel (this is the fragment separation gel, in short
the "sequencing gel") having gaps of a known average size. Migration
occurs because of the strong electric charge of the fragments (DNA is a
highly charged molecule, with a charge directly proportional to the number
of bases it comprises). Practically, one loads a small volume of the
mixture at the top of a gel on which is applied an electric field. The
different fragments will then move snake-like by reptation inside the gel.
And one easily understands that the shorter fragments having less
difficulty to go through the gel pores will move faster than the longer
ones. One will therefore obtain after some time a ladder separating
fragments according to their exact length. The subsequent step will be to
identify the place where they are located, to place them with respect to
each other.
In the first experiments one used a process — which is still in use today
— to label either the primer used for polymerization, or the nucleotides
as they are incorporated into the newly synthesized fragment, with a
radioactive isotope. As a consequence, the fragments in the gel were
radioactive. It was therefore easy to reveal their presence by placing the
gel on a photograph plate and detecting the black silver bands in the
resulting photograph. To allow detection of all four nucleotides this
experiment asked for the running of four separate mixtures coming from
four different assays in different test tubes in different lanes in a gel.
One thus obtained four mixtures where the fragments were ending at each of
the nucleotides A, T, G and C. The mixture then ran side by side. After
photograph development, this gives rise to four sequences of black bands
where one can directly read the sequence. It is this image of a ladder
made of parallel black bands that was interpreted by many journalists as
the (rather misleading and difficult to understand) metaphor of a "bar
code" as representing fragments of chromosomes. In a standard experiments,
for gels 50 centimeters long, one reads 500 bases, with an error rate of
about 1%. This has as a consequence that it is usually difficult, and also
long and costly, to determine the sequence of a long fragment of DNA with
an error rate lower than one in ten thousand.
Of course labeling can be performed using fluorescent
chemicals, and detection of the bands uses lasers and light detectors.
Assembly
The "shotgun" sequencing step produces thousands of
sequences which, once correctly assembled, make up the whole original
sequence. Suitable algorithms compare the sequences in pairs, looking for
the best possible alignment, then identifying overlapping regions and
matching them to reconstruct the original DNA. Early programs, developed
at a time when computers were not powerful enough to envisage assembling
large numbers of sequences, used algorithms which compared sequences in
pairs. However, random sequencing of large genomes involves assembling not
just thousands of sequences but tens of thousands, even millions, and with
the need to handle these increasing numbers of sequences, it became clear
that the methods available were not specialized enough. A point was
reached where the computational side was holding sequencing projects back,
and the biologist had to make up for the inadequacy of the processing by
spending time “helping” the assembly programs. In certain cases
reconstituting the whole text from the text of fragments can be not merely
difficult but even impossible, particularly when the text includes long
repeated regions, a common occurrence with long segments or whole genomes.
All this led several laboratories to develop new algorithms, bringing
together the biologist’s knowledge, information about the nature of each
of the fragments to be sequenced, (such as the sequencing strategy used
for a given segment of DNA, the source, and the approximate length), and
pairwise or multiple alignment algorithms. As a final stage, so that users
can verify the result of the assembly procedure and spot possible errors
or inconsistencies, graphic interfaces are used to view and correct the
assembly if necessary. This is done either as a whole, by comparing the
different segments, or locally, by correcting the raw results of
sequencing each fragment. However the aim of using computational methods
is to reduce the user’s intervention to a minimum, producing a correct
final result directly. An enormous amount of work has been put into this
since the middle of the 1990s, and it was crowned with success when the
first draft sequence of the human genome was completed in the year 2000.
 DNA Chips and Transcriptome Analysis
DNA Chips and Transcriptome Analysis
The interest for DNA chips is that they provide large scale
information on thousands of genes at the same time. This very fact implies
some sort of statistical reasoning when studying the intensities of the
spots analyzed.
1. What kind of intensity distribution should be expected?
A general theorem of statistics states that the outcome of a series of
multiple additive causes should result in a Laplace-Gauss distribution of
intensities. Clearly, transcription is not additive, but, rather,
multiplicative (each mRNA level comes from the multiplication of many
causes, from the metabolism of individual nucleotides to the
compartmentalization in the cell). One expects therefore that the
logarithm of the intensities should display a Laplace-Gauss distribution
("log-normal" distribution). This is indeed what is observed. A
first criterion of validity of an experiment with DNA arrays is therefore
to check that the distribution is log-normal. In fact the situation is
more complicated (and more interesting) and deviations from the
Laplace-Gauss log-normal situation can be exploited to uncover unexpected
links between genes that are expressed together. This is the case, in
particular, when sets of genes behave independently from each
other.
2. Many causes can affect transcription efficiency. This
has the consequence that hybridization on DNA chips is highly sensitive to
experimental errors. It is therefore necessary to have a significant
number of control experiments, which may identify some of the most
frequent experimental errors. It should be noted that these errors are
biologically significant and tell something on the organism of interest.
It is therefore of the utmost importance to try to differentiate, in a
given experiment, what pertains to this kind of systematic error, and what
pertains to the phenomenon under investigation. An example of such study
is presented in an
article from the HKU-Pasteur Research Centre.
A general library of references on the use of DNA chips is
provided here.
Work on the ubiquitous regulator H-NS illustrates how
transcriptome analysis may be used to better understand the role of a
regulator located high in the control hierarchy of gene expression in
bacteria.
 2D Electrophoresis Gels and Proteome Analysis
2D Electrophoresis Gels and Proteome Analysis
|
pI = 4 Separation by electric charge pI =
7
|
|
| The cell is broken and its protein extracted in a soluble form.
In a first step they are separated according to their electric
charge (isoelectric point, pI). Then the first dimension gel is
deposited horizontally on top of a mesh that will separate the
proteins according to their molecular mass
Finally the proteins are stained using silver nitrate. They
appear as black spots, more or less intense, according to the
total amount of the protein present in the gel.
Proteins are identified by various methods, mass spectrometry
in particular.
|
 |
M
o
l
e
c
u
l
a
r
M
a
s
s
|
|
|
|
The total set of the proteins in a cell is named its
"proteome" (a word build up in the same way as "genome"). 2D gel
electrophoresis is a quantitative way to display the part of the
proteome of a cell that is expressed under particular conditions.
Usually, most of the proteins displayed are in a narrow range (slightly
acidic pI = 4 to pI = 7) and for molecular masses higher than 10
kilodaltons (i.e. more than about 100 amino acids). Small proteins, and
basic or very acidic proteins are difficult to visualize with this
technique. Moreover, for the time being, it is still difficult to
extract and separate in a reproducible way the proteins that are
imbedded in biological membranes. Nevertheless proteome analysis by 2D
gel electrophoresis is still extremely important. Not only does this
technique give the final state of gene expression in a cell, but it also
displays proteins as they are found, i.e. after they have been modified
post-translationally.
Work on the ubiquitous regulator H-NS illustrates how
proteome analysis may be used to better understand the role of a
regulator located high in the control hierarchy of gene expression in
bacteria.
 Metabolism
Metabolism
Living organisms continuously transform matter chemically.
As a matter of fact, this is a way to ensure that a system is alive. A
spore, or a seed, will only be considered as alive when it germinates,
i.e. at a moment when it will transform some molecules into other
molecules. This is the basis of metabolism. There is an intermediate
stage, between life and death, named "dormancy", but this is only
understood to be a true state after the fact, that is, after the moment
when some metabolic property has been witnessed.
 In
silico Genome Analysis (BioInformatics)
In
silico Genome Analysis (BioInformatics)
 Some Major Genomics Sites and Supporting
Agencies
Some Major Genomics Sites and Supporting
Agencies
 The US Department of Energy
The US Department of Energy
 The
US National Institutes of Health
The
US National Institutes of Health
 The
European Union Directorate General XII
The
European Union Directorate General XII
 Japan
Science and Technology Corporation ( JST)
Japan
Science and Technology Corporation ( JST)
 The Sanger Centre
The Sanger Centre
 The Kasuza DNA Research
Institute
The Kasuza DNA Research
Institute
 Washington University
Washington University
 Oklahoma University
Oklahoma University
 Baylor College
Baylor College
 The Whitehead Institute
The Whitehead Institute
 The Institute for Genome Research
(TIGR)
The Institute for Genome Research
(TIGR)
 Le Centre National de Séquençage
(Genoscope)
Le Centre National de Séquençage
(Genoscope)
 The Beijing Human Genome Centre
The Beijing Human Genome Centre
 The Shanghai Genome Centre
The Shanghai Genome Centre
 The International
Nucleotide Sequences Database Collaboration
The International
Nucleotide Sequences Database Collaboration
 The Reference Protein Data Bank
at Expasy (SwissProt) (mirror in
Beijing)
The Reference Protein Data Bank
at Expasy (SwissProt) (mirror in
Beijing)
![]() A Brief History of the Human Genome
Initiative
A Brief History of the Human Genome
Initiative ![]() DNA Chips and Transcriptome Analysis
DNA Chips and Transcriptome Analysis
![]() 2D Electrophoresis Gels and Proteome Analysis
2D Electrophoresis Gels and Proteome Analysis ![]() Metabolism
Metabolism ![]() In silico Genome Analysis (BioInformatics)
In silico Genome Analysis (BioInformatics)