Research
I am currently a postdoc in the Lifeware team at Inria Saclay, advised by Jakob Ruess, and I am working on stochastic models of cell populations.
Previously, I was a postdoc in Mary Dunlop’s lab at Boston University.
I am passionate about mathematical modelling and scientific programming, which is what underlines my past and current research.
All links below correspond to publications that I authored or co-authored, some of them belonging in several categories: software, journal article, preprint, thesis, conference paper.
Current research interests
Bayesian statistics and causal inference
Learning about bayesian statistics and causal inference changed my way to approach research problems. As a core developer of PyMC, the leading Python library for bayesian inference, I was involved in the release of version 5 of the library (PeerJ CS 2023).
This framework was instrumental to solve an inverse problem in combinatorial protein design with Nathan Tague (ACS Synth. Biol. 2023). Explicit consideration of the randomness and noise sources in the experimental process enabled us to build a white-box model that was able to extract the relevant information from the data, while classical approaches could not.
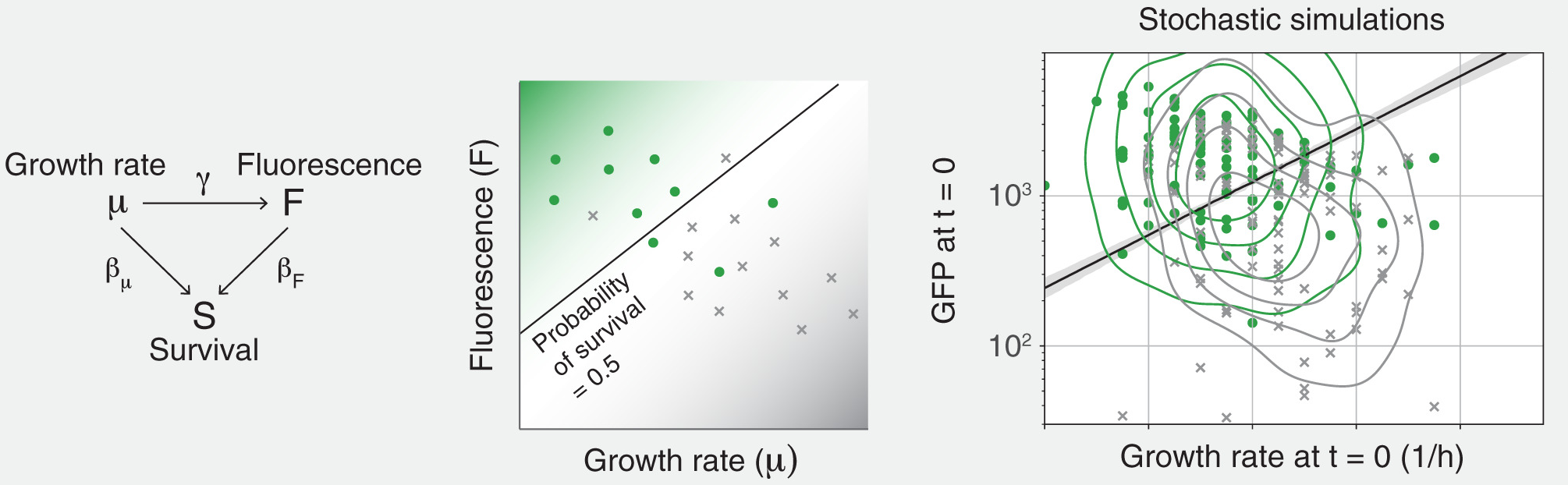
I also used it to investigate the heterogeneity of cell response to antibacterial treatments, with Nadia Sampaio and Caroline Blassick (PNAS 2022). Careful causal modeling enabled us to determine the role of stress pathways in the survival of bacterial cells to antibiotics.
As a continuation of this project, with Razan Alnahhas, we worked on deciphering the interactions of stress pathways in cell survival, with dual-reporter strains (mSystems 2025).
Scientific programming
I have always been fascinated with computers and computational challenges. I really enjoy programming, and I prefer developing clean, useful and reusable libraries than throwaway research scripts.
For almost the last three years, I have been the principal maintainer of DeLTA, our deep neural network segmentation and tracking software for microscopy images. I have been overhauling it in view of a version 3 release, to come.
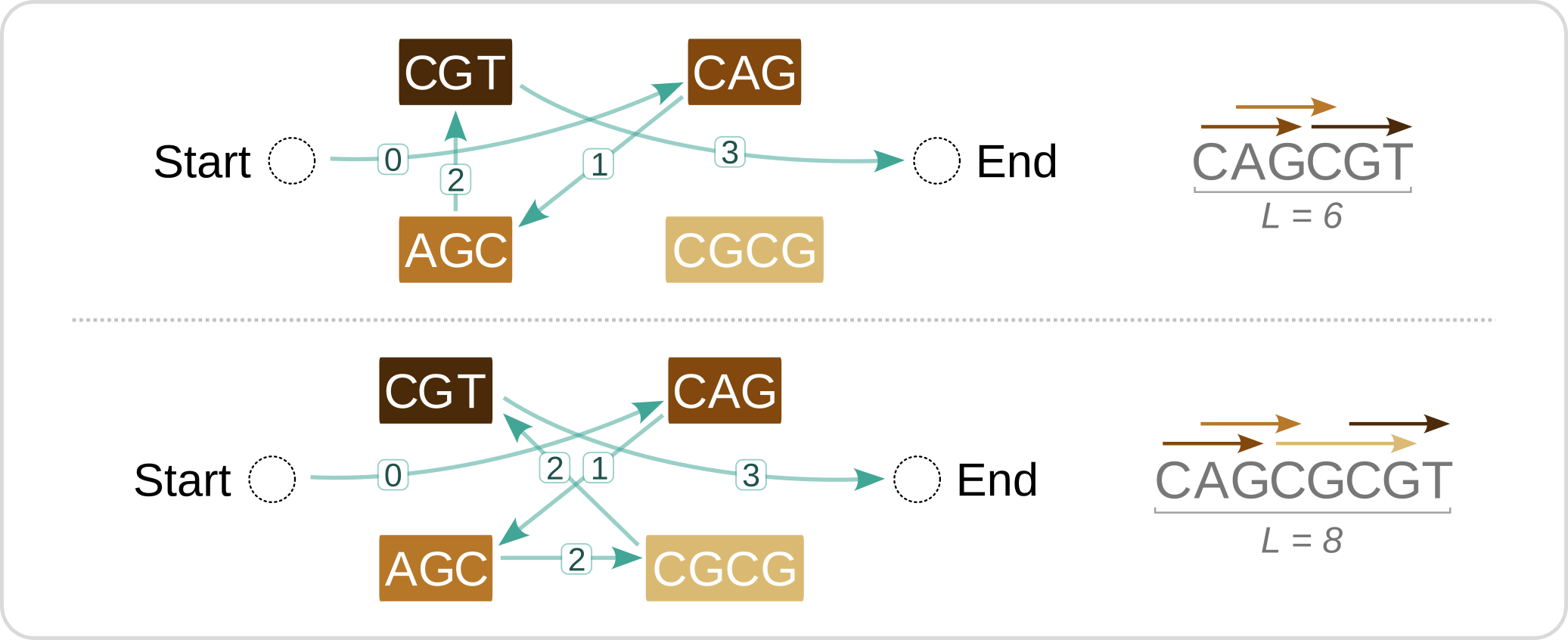
With Eric South, I tackled the NP-hard problem of promoter design by devising an integer linear programming approach inspired by the Traveling Salesman Problem (PLOS CB 2024). Our open-source implementation is available as the Python library dense-arrays.
During my thesis, with Élise Weill, I used optimal control of a system of differential equations to optimize bioproduction (ECC 2019).
I am the author of rebop, one of the fastest libraries to simulate stochastic reaction networks.
Finally, my interests in hacking and electronics allowed me to reverse-engineer a plate reader communication protocol and write a Python USB driver for it, platerider, to extend the plate reader’s programmatic features.
Antibiotic resistance, resilience and tolerance
Bacteria, while being some of the simple organisms we know, can already exhibit complex behaviors, even more so when considered as part of a population.
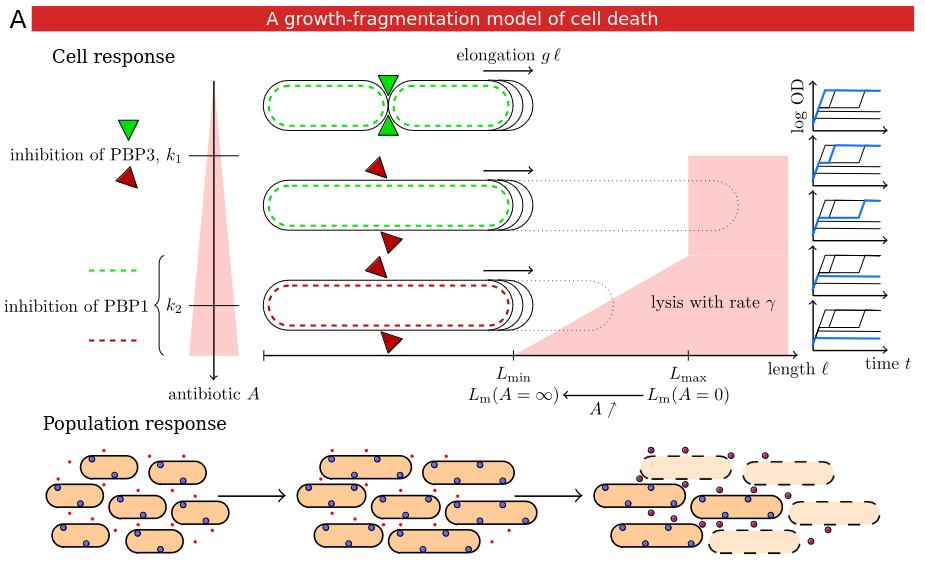
During the course of my PhD thesis (IP Paris 2020), I initially helped Hannah Meredith define the notion of resilience of a bacterial population to an antibiotic treatment (Science Advances 2018). Then, I developed a multiscale mathematical model of the response of a cell population to an antibiotic treatment, enabling treatment optimization (bioRxiv 2021). I had a small role in a later study by Kyeri Kim and coauthors investigating the mechanism of filamented cell lysis (Mol Syst Biol 2023), one of the key mechanisms of my model.
Lately, I was using single-cell data from microfluidic images to investigate the interactions between stress pathways and their causal role in the survival of bacteria to antibiotic treatments (PNAS 2022 with Nadia Sampaio and Caroline Blassick, and with Razan Alnahhas on a continuation of this work with dual-reporter strains mSystems 2025).
Education
- I studied at École Normale Supérieure from 2011 to 2015
- I got a Masters degree in Quantum Physics in 2014
- And a Masters degree in Theoretical Computer Science in 2015
- I then worked on my PhD in Computational Biology from 2016 to 2020 at Inria, École polytechnique and Institut Pasteur under the supervision of Gregory Batt