Deep learning (DL) methods show great success in handling sequential data, including text (natural language processing) and human movement (action prediction). However, they are scarcely used to understand animal behavior. However, motion carries rich additional information about neuronal and motor functioning. Animal models, such as rodents, can be studied under different medical conditions and in different environments. With recent advances in cameras and computer vision, animal behaviour can be measured through video recordings and motion capture systems, producing high quality data of an individual’s motion. The dynamics of postures that generate behaviours are complex, spanning multiple spatio-temporal scales, and there is currently a lack of computational methods to tackle this complexity.
In this project, I develop new DL methods for quantitative description of animal movement, more particularly to better understand how normal behaviors are impaired under specific pharmacological treatments. They will lead to better understand how normal behaviors (e.g. walking, balance, orientation in space, very subtle motion changes) are impaired under specific pharmacological treatments. This project also aims at improving deep learning methods, by comparing, testing and combining different modularities of neural networks, focusing on unsupervised learning, and pushing the understanding of inferred representations. The resulting methods will be applicable to other domains and most importantly can pave the way to detailed quantitative study of human behaviour including its early changes preceding neurodegenerative deterioration of the brain. Decoding behaviour - finding out what it means and predicting it - bears a great potential for improved diagnostics and new therapeutic strategies for neural disorders.
Keywords: behavior, representation learning, recurrent neural networks (RNNs), graph convolutional neural networks (GCNs), transformers, missing data, data imputation, in-motion neural recordings
Imputing missing data from 2D and 3D keypoint traking
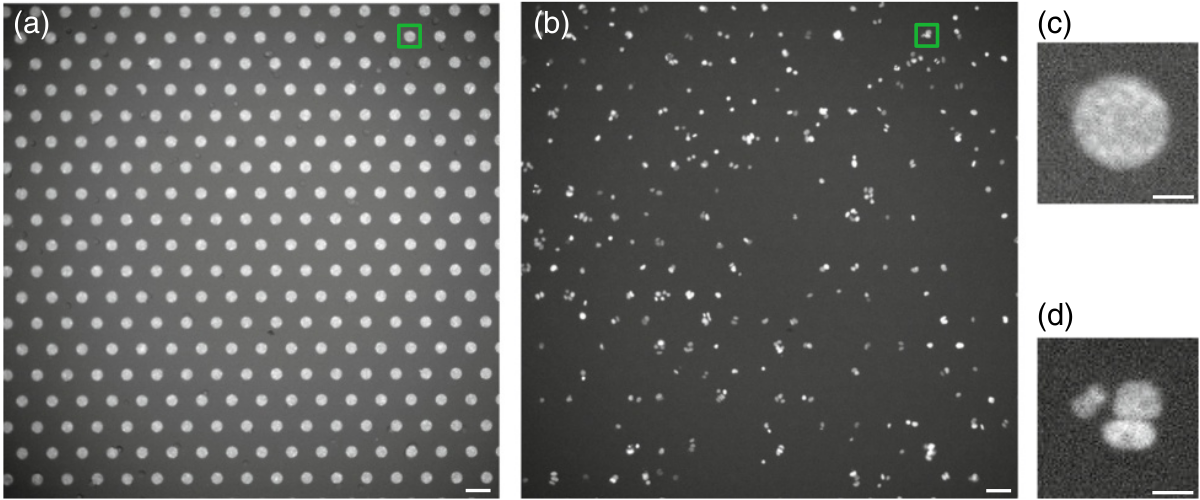
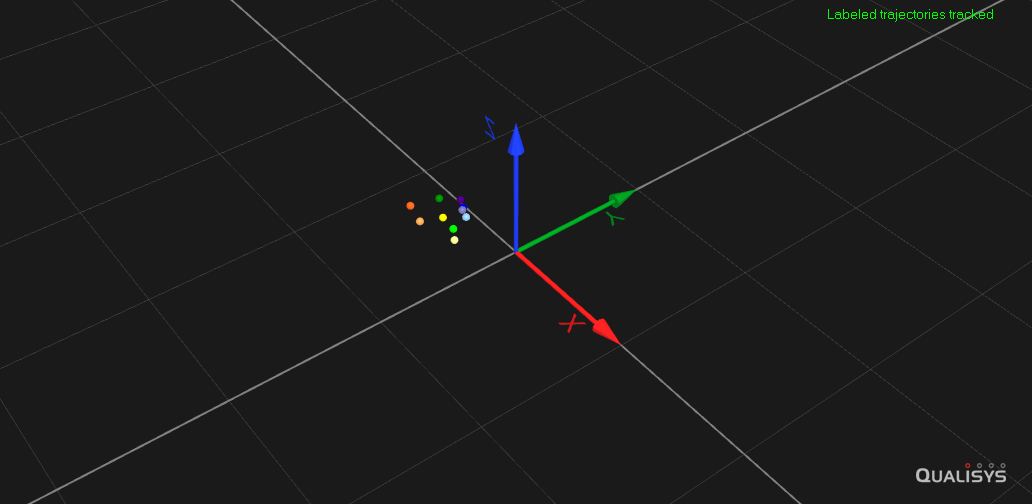
Both video recordings coupled with automated posture estimation and motion capture systems are producing high quality data of an individual’s motion. Yet these methods are not perfect and contain missing data. I developed Deep Imputation for Skeleton data (DISK), a deep learning algorithm to learn dependencies between keypoints and their dynamics to impute missing tracking data. We demonstrated the usability and performance of our imputation method on six different animal skeletons including two multi-animal set-ups.
Using DISK, I am exploring behavior representations and detecting automatically drug-induced differences in freely behaving mouse 3D recordings performing locomotory tasks like floor exploration, climbing, or running on a fixed-speed treadmill.
- Here is the poster I presented at the Society for Neuroscience 2024 conference in Chicago.
- Here is the talk I gave at the Machine Learning MLinPL conference in 2024 in Warsaw.
- The code is available on github .
- A tutorial is available as a jupyter notebook hosted on google colab.
- The University of Cologne and scisimple are talking about DISK!
Other resources: